Reanalysis of ribosome profiling datasets reveals a novel function of rocaglamide A in perturbing the dynamics of translation elongation via eIF4A
Fajin Li1,2,3,4, Jianhuo Fang1,4, Yifan Yu1,4, Sijia Hao1,2,3, Qinglin Zeng1, Qin Zou1, and Xuerui Yang1,2,3
1 MOE Key Laboratory of Bioinformatics, School of Life Sciences, Tsinghua University, Beijing 100084, China 2 Center for Synthetic & Systems Biology, Tsinghua University, Beijing 100084, China 3 Joint Graduate Program of Peking-Tsinghua-National Institute of Biological Science, Tsinghua University, Beijing 100084, China. 4 These authors are main contributors to this work.
Corresponding: yangxuerui@tsinghua.edu.cn; lfj17@tsinghua.org.cn; sherkinglee@gmail.com
This file is a description of how the results presented in the manuscript were generated, including the datasets we used, how we downloaded and processed the raw datasets, the codes we used, et al. All scripts used for processing BAM files should be excuted in Linux platform and other scripts can be both used in Linux and windows platform. It should be noted that most of the analyses were based on RiboMiner we developed before.
The datasets we used were all downloaded from GEO database. We all collected 26 datasets, containing more than 100 ribosome profiling samples and several iCLIP-seq samples. Please refer to Table S1 for the detailed information of all the collected datasets.
The codes used for downloading all the datasets are:
# use GSE102720 as an example:
$ cat download.sh
workdir=/workdata/LabMember2/lifj/lifj/Project/05.Ribo_seq_human/GSE102720/00.rawdata
for i in SRR59376{40..45};do
fastq-dump $i
rm /workdata/LabMember2/lifj/ncbi/public/sra/$i.sra
done
- python >= 3.6
- RiboMiner = 0.2.3.2
- snakemake = 5.31.1
- sra-tools = 2.11.0
- fastx_toolkit = 0.0.14
- cutadapt = 4.0
- bowtie >= 1.1.2
- STAR = 2.7.7a
- samtools = 1.11
- R >= 3.6
The human reference genome, ncRNA and annotations are downloaded from Ensemble Genome Browser (release 88). Human rRNA sequences are downloaded from UCSC Genome Browser and download method could refer to this protocol.
- Download and install some tools and packages
# python >=3.6
conda install -c bioconda ribocode ribominer sra-tools
fastx_toolkit cutadapt bowtie star samtools snakemake
- Build index for rRNA alignment:
fastaGenome=hg38_rRNA.fa
bowtie-build -f $fastaGenome human_rRNA
- Build index for genome alignment:
fastaGenome=Homo_sapiens.GRCh38.dna.primary_assembly.fa
gtf=Homo_sapiens.GRCh38.88.gtf
srun STAR --runThreadN 8 --runMode genomeGenerate --genomeDir ./STAR_Human_Ensembl_GRCh38_Ensembl --genomeFastaFiles $fastaGenome --sjdbGTFfile $gtf
- Get annotations for transcriptome
prepare_transcripts -g <Homo_sapiens.GRCh38.88.gtf> -f <Homo_sapiens.GRCh38.dna.primary_assembly.fa> -o <RiboCode_annot>
OutputTranscriptInfo -c <RiboCode_annot/transcripts_cds.txt> -g <Homo_sapiens.GRCh38.88.gtf> -f <RiboCode_annot/transcripts_sequence.fa> -o <longest.transcripts.info.txt> -O <all.transcripts.info.txt>
All ribosome profiling and Disome-seq datasets were processed with a custom snakemake pipeline. Use GSE102720 as an example,run the snakemake file:
snakemake -s Ribo-seq-snakemake.py --cluster "sbatch -p cmp -N 2 -n 8 -e Ribo.err -o Ribo.out " --jobs 61
Before doing the following analyses, all datasets we collected should be processed with the Ribo-seq-pipeline.
- Set configure files (GSE102720 for example)
bamFiles readLengths Offsets bamLegends
../07.STAR/SRR5937640_STAR/SRR5937640.Aligned.toTranscriptome.out.sorted.bam 26,27,28,29,30,31 12,12,12,12,12,13 Naive-DMSO-1
../07.STAR/SRR5937641_STAR/SRR5937641.Aligned.toTranscriptome.out.sorted.bam 26,27,28,29,30,31 12,12,12,12,12,13 Naive-DMSO-2
../07.STAR/SRR5937642_STAR/SRR5937642.Aligned.toTranscriptome.out.sorted.bam 26,27,28,29,30,31 12,12,12,12,12,13 Naive-RocA03
../07.STAR/SRR5937643_STAR/SRR5937643.Aligned.toTranscriptome.out.sorted.bam 26,27,28,29,30,31 12,12,12,12,12,13 Aglaiacized-DMSO-1
../07.STAR/SRR5937644_STAR/SRR5937644.Aligned.toTranscriptome.out.sorted.bam 26,27,28,29,30,31 12,12,12,12,12,13 Aglaiacized-DMSO-2
../07.STAR/SRR5937645_STAR/SRR5937645.Aligned.toTranscriptome.out.sorted.bam 26,27,28,29,30,31 12,12,12,12,12,13 Aglaiacized-RocA03
- Calculate polarity scores using RiboMiner
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA
attribute=$workdir/GSE102720_configure.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='Naive-DMSO,Naive-RocA03,Aglaiacized-DMSO,Aglaiacized-RocA03'
replicates='Naive-DMSO-1,Naive-DMSO-2__Naive-RocA03__Aglaiacized-DMSO-1,Aglaiacized-DMSO-2__Aglaiacized-RocA03'
PolarityCalculation -f $attribute -c $trans_info -o $results/MA_all -n 0
PlotPolarity -i $results/MA_all_polarity_dataframe.txt -o $results/MA_all -g $groups -r $replicates -y 5 --mode all
- Calculate the difference of polarity scores
import pandas as pd
df=pd.read_csv('MA_all_mean_polarity_dataframe.txt',sep="\t")
print(df.shape)
##19636,5
m,n=df.shape
print(df.iloc[0:5,0:(n-1)])
print(df.columns)
df.rename(columns={'Unnamed: 0':'transcript_id'}, inplace = True)
print(df.columns)
df['Naive-RocA03-DMSO(GSE102720)']=df['Naive-RocA03']-df['Naive-DMSO']
df['Aglaiacized-RocA03-DMSO(GSE102720)']=df['Aglaiacized-RocA03']-df['Aglaiacized-DMSO']
df_new=df[['transcript_id','Naive-RocA03-DMSO(GSE102720)','Aglaiacized-RocA03-DMSO(GSE102720)']]
print(df_new.shape)
print(df_new.iloc[0:5,:])
df_new.to_csv('GSE102720_diff_polarity_dataframe.txt',sep="\t",index=False)
Finally, the difference of polarity scores from all datasets were merged. Please refer to total_merged_diff_polarity_202102.txt.
- use GSE102720 for example.
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA
attribute=$workdir/GSE102720_configure.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='Naive-DMSO,Naive-RocA03,Aglaiacized-DMSO,Aglaiacized-RocA03'
replicates='Naive-DMSO-1,Naive-DMSO-2__Naive-RocA03__Aglaiacized-DMSO-1,Aglaiacized-DMSO-2__Aglaiacized-RocA03'
mkdir -p $workdir/RPKM
python RPKM.py -f $attribute -c $trans_info -o $workdir/RPKM/MA --type CDS ## calculate RPKM of each longest transcript
python RPKMmean.py -i $workdir/RPKM/MA_RPKM_dataframe.txt -o $workdir/RPKM/MA -g $groups -r $replicates ## calculate mean RPKM for the replicates
- Calculate the log2FC(RPKM) for each transcript
import pandas as pd
import numpy as np
df=pd.read_csv('MA_mean_RPKM_dataframe.txt',sep="\t")
print(df.shape)
##19636,5
m,n=df.shape
print(df.iloc[0:5,0:(n-1)])
print(df.columns)
df.rename(columns={'Unnamed: 0':'transcript_id'}, inplace = True)
print(df.columns)
df['Naive-RocA03-DMSO(GSE102720)']=np.log2(df['Naive-RocA03']/df['Naive-DMSO'])
df['Aglaiacized-RocA03-DMSO(GSE102720)']=np.log2(df['Aglaiacized-RocA03']/df['Aglaiacized-DMSO'])
df_new=df[['transcript_id','Naive-RocA03-DMSO(GSE102720)','Aglaiacized-RocA03-DMSO(GSE102720)']]
print(df_new.shape)
print(df_new.iloc[0:5,:])
df_new.to_csv('GSE102720_diff_RPKM_dataframe.txt',sep="\t",index=False)
Finally, the RPKM of all transcripts from all datasets were merged. Please refer to total_merged_cds_level_RPKM_202102.txt.
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA
attribute=$workdir/GSE102720_configure.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='Naive-DMSO,Naive-RocA03,Aglaiacized-DMSO,Aglaiacized-RocA03'
replicates='Naive-DMSO-1,Naive-DMSO-2__Naive-RocA03__Aglaiacized-DMSO-1,Aglaiacized-DMSO-2__Aglaiacized-RocA03'
## coverage
mkdir -p $workdir/coverage
RiboDensityAtEachPosition -c $trans_info -f $attribute -o $workdir/coverage/MA -U codon
Finally, the coverage of all transcripts from all datasets were merged. Please refer to total_merged_read_coverage_202102.txt.
- all used data were deposited in ./data
TranslationRelatedConsitionPairs.txt
total_merged_cds_level_RPKM_202102.txt
total_merged_diff_polarity_202102.txt
total_merged_read_coverage_202102.txt
- clustering analyses via RiboPipe.py
#!/usr/bin/env python
# -*- coding:UTF-8 -*-
'''
Author: Li Fajin
Date: 2021-02-15 13:23:59
LastEditors: Li Fajin
LastEditTime: 2022-05-28 12:21:37
Description: file content
'''
import pandas as pd
import numpy as np
import matplotlib
matplotlib.use('Agg')
import matplotlib.pyplot as plt
import seaborn as sns
import os
'''
###################################
Date: 20220528.15:00
all polarity with Tranlation initiation and elongation
19636 trans x 49 conditionPairs
###################################
'''
ConditionPairs_of_initiation_and_elongation=pd.read_csv("../data/TranslationRelatedConsitionPairs.txt",sep="\t")
polarity_with_initiation_and_elongation=pd.read_csv("../data/total_merged_diff_polarity_202102.txt",sep="\t",index_col=0)
RPKM_with_initiation_and_elongation=pd.read_csv("../data/total_merged_cds_level_RPKM_202102.txt",sep="\t",index_col=0)
RPKM_with_initiation_and_elongation.apply(np.mean, axis=1).describe() ## trans describe
RPKM_with_initiation_and_elongation.apply(np.mean, axis=0).describe() ## sample describe
coverage_with_initiation_and_elongation=pd.read_csv("../data/total_merged_read_coverage_202102.txt",sep="\t",index_col=0)
coverage_with_initiation_and_elongation_sorted=coverage_with_initiation_and_elongation[sorted(coverage_with_initiation_and_elongation.columns)]
coverage_with_initiation_and_elongation_sorted.apply(np.mean, axis=1).describe() ## trans describe
coverage_with_initiation_and_elongation_sorted.apply(np.mean, axis=0).describe() ## sample describe
RPKM_with_initiation_and_elongation_filtered_gt0_in_each_samples=RPKM_with_initiation_and_elongation.loc[RPKM_with_initiation_and_elongation.apply(np.min,axis=1)>=1,:]
coverage_with_initiation_and_elongation_filtered_mean_of_each_trans_gt_pectMean=coverage_with_initiation_and_elongation_sorted.loc[coverage_with_initiation_and_elongation_sorted.apply(np.mean,axis=1)>=coverage_with_initiation_and_elongation_sorted.apply(np.mean,axis=1).describe()['mean'],:]
trans_in_common_associated_with_translation=set(RPKM_with_initiation_and_elongation_filtered_gt0_in_each_samples.index).intersection(set(coverage_with_initiation_and_elongation_filtered_mean_of_each_trans_gt_pectMean.index))
polarity_with_initiation_and_elongation_filtered=polarity_with_initiation_and_elongation.loc[trans_in_common_associated_with_translation,:]
polarity_with_initiation_and_elongation_filtered_vars=np.var(polarity_with_initiation_and_elongation_filtered,axis=1)
polarity_with_initiation_and_elongation_filtered_vars.describe()
trans_filtered_by_polarityVars=polarity_with_initiation_and_elongation_filtered.iloc[np.where(polarity_with_initiation_and_elongation_filtered_vars>polarity_with_initiation_and_elongation_filtered_vars.describe()[1]+polarity_with_initiation_and_elongation_filtered_vars.describe()[2])[0],:]
polarity_with_initiation_and_elongation_filtered_filtered_byPolarityVars=polarity_with_initiation_and_elongation_filtered.drop(trans_filtered_by_polarityVars.index)
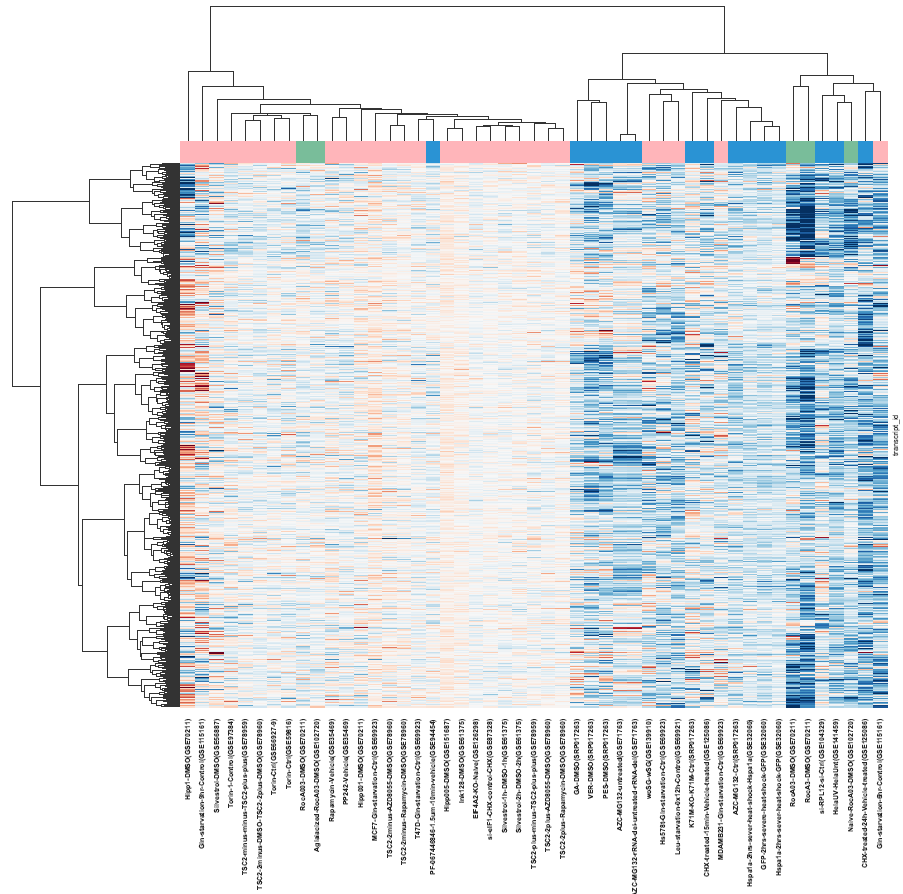
plt.rc('font',weight='bold')
sns.set(font_scale=0.5)
fig=plt.figure()
col_c=dict(zip(ConditionPairs_of_initiation_and_elongation['TranslationType'].unique(), ['#2a93d4','#ffb5ba','#79bd9a']))
col_colors=ConditionPairs_of_initiation_and_elongation['TranslationType'].map(col_c)
ax=sns.clustermap(polarity_with_initiation_and_elongation_filtered_filtered_byPolarityVars,method ='ward',metric='euclidean',row_cluster=True,col_cluster=True,
vmin=-0.5,vmax=0.5,center=0,cmap = 'RdBu_r',col_colors=col_colors.values,yticklabels=False,xticklabels=True)
ax.cax.set_visible(False)
plt.savefig("../results/Figure 1A.pdf")
- set configure file
bamFiles readLengths Offsets bamLegends
../07.STAR/SRR2075925_STAR/SRR2075925.Aligned.toTranscriptome.out.sorted.bam 25,26,27,28 11,11,11,11 DMSO-1
../07.STAR/SRR2075926_STAR/SRR2075926.Aligned.toTranscriptome.out.sorted.bam 25,26,27,28,29 11,11,11,11,12 DMSO-2
../07.STAR/SRR2075927_STAR/SRR2075927.Aligned.toTranscriptome.out.sorted.bam 25,26,27,28,29 11,11,11,11,12 RocA-003
../07.STAR/SRR2075928_STAR/SRR2075928.Aligned.toTranscriptome.out.sorted.bam 24,25,26,27,28,29 11,11,11,11,11,11 RocA-03
../07.STAR/SRR2075929_STAR/SRR2075929.Aligned.toTranscriptome.out.sorted.bam 26,27,28,29 11,11,12,12 RocA-3
All the bam files were generated via the Ribo-seq analyses pipeline.
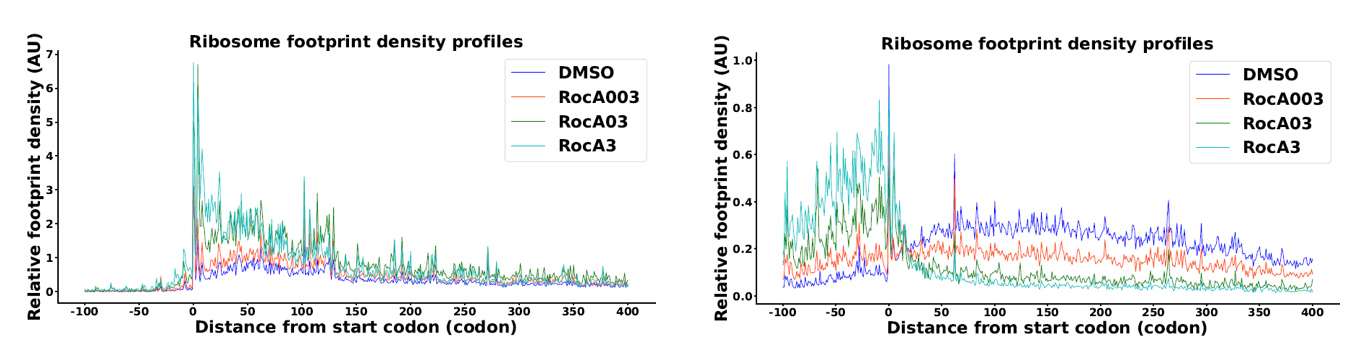
- Metagene via RiboMiner
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA
attribute=$workdir/GSE70211_configure.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='DMSO,RocA003,RocA03,RocA3'
replicates='DMSO-1,DMSO-2__RocA-003__RocA-03__RocA-3'
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA -U codon -M RPKM -u 100 -d 400 -l 100 -n 10 -m 1 --norm no -y 100 --CI 0.95 --type UTR
PlotMetageneAnalysis -i $results/MA_unnormed_dataframe.txt -o $results/MA_unnormed -g $groups -r $replicates -u 100 -d 400 --mode mean
- Calculate codon density for each sample
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA
attribute=$workdir/GSE70211_configure.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='DMSO,RocA003,RocA03,RocA3'
replicates='DMSO-1,DMSO-2__RocA-003__RocA-03__RocA-3'
RiboDensityAtEachPosition -c $trans_info -f $attribute -o $workdir/coverage/MA -U codon
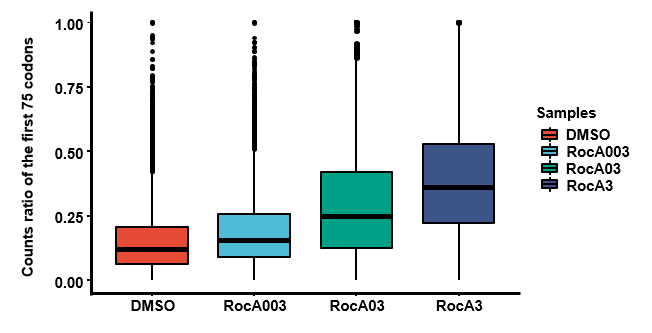
- Statistic read density at the first 75 codons via ProcessCodonDensityAtEachPosition.py script.
## the first 75
$ python ProcessCodonDensityAtEachPosition.py -i MA_DMSO-1_cds_codon_density.txt -l 1 -r 75 -o DMSO_1_first75.txt
$ python ProcessCodonDensityAtEachPosition.py -i MA_DMSO-2_cds_codon_density.txt -l 1 -r 75 -o DMSO_2_first75.txt
$ python ProcessCodonDensityAtEachPosition.py -i MA_RocA-003_cds_codon_density.txt -l 1 -r 75 -o RocA003_first75.txt
$ python ProcessCodonDensityAtEachPosition.py -i MA_RocA-03_cds_codon_density.txt -l 1 -r 75 -o RocA03_first75.txt
$ python ProcessCodonDensityAtEachPosition.py -i MA_RocA-3_cds_codon_density.txt -l 1 -r 75 -o RocA3_first75.txt
## all CDS
$ python ProcessCodonDensityAtEachPosition.py -i MA_DMSO-1_cds_codon_density.txt -o DMSO_1_all.txt
$ python ProcessCodonDensityAtEachPosition.py -i MA_DMSO-2_cds_codon_density.txt -o DMSO_2_all.txt
$ python ProcessCodonDensityAtEachPosition.py -i MA_RocA-003_cds_codon_density.txt -o RocA003_all.txt
$ python ProcessCodonDensityAtEachPosition.py -i MA_RocA-03_cds_codon_density.txt -o RocA03_all.txt
$ python ProcessCodonDensityAtEachPosition.py -i MA_RocA-3_cds_codon_density.txt -o RocA3_all.txt
- Statistics of ratio of the first 75 codons vs the whole CDS region,via RiboDensityAtEachPosition.Rmd.
## Open Rstudio and do it in R platform
rm(list=ls())
setwd('../data')
human_info <- read.table("Homo_sapiens.GRCh38.88.transInfo.txt",sep="\t",stringsAsFactors = F,header = T,check.names = F)
## all counts
DMSO_1_all_counts <- read.table("DMSO_1_all.txt",sep="\t",header = T,stringsAsFactors = F,row.names = 1)
DMSO_2_all_counts <- read.table("DMSO_2_all.txt",sep="\t",header = T,stringsAsFactors = F,row.names = 1)
RocA003_all_counts <- read.table("RocA003_all.txt",sep="\t",header = T,stringsAsFactors = F,row.names = 1)
RocA03_all_counts <- read.table("RocA03_all.txt",sep="\t",header = T,stringsAsFactors = F,row.names = 1)
RocA3_all_counts <- read.table("RocA3_all.txt",sep="\t",header = T,stringsAsFactors = F,row.names = 1)
## first 75 codons
DMSO_1_first75 <- read.table("DMSO_1_first75.txt",sep="\t",header = T,stringsAsFactors = F,row.names = 1)
DMSO_2_first75 <- read.table("DMSO_2_first75.txt",sep="\t",header = T,stringsAsFactors = F,row.names = 1)
RocA003_first75 <- read.table("RocA003_first75.txt",sep="\t",header = T,stringsAsFactors = F,row.names = 1)
RocA03_first75 <- read.table("RocA03_first75.txt",sep="\t",header = T,stringsAsFactors = F,row.names = 1)
RocA3_first75 <- read.table("RocA3_first75.txt",sep="\t",header = T,stringsAsFactors = F,row.names = 1)
## all counts
DMSO_1_factor <- sum(DMSO_1_all_counts$counts)
DMSO_2_factor <- sum(DMSO_2_all_counts$counts)
RocA003_factor <- sum(RocA003_all_counts$counts)
RocA03_factor <- sum(RocA03_all_counts$counts)
RocA3_factor <- sum(RocA3_all_counts$counts)
## normalized
DMSO_1_all_counts['RPM'] <- 10^6*(DMSO_1_all_counts/DMSO_1_factor)
DMSO_2_all_counts['RPM'] <- 10^6*(DMSO_2_all_counts/DMSO_2_factor)
RocA003_all_counts['RPM'] <- 10^6*(RocA003_all_counts/RocA003_factor)
RocA03_all_counts['RPM'] <- 10^6*(RocA03_all_counts/RocA03_factor)
RocA3_all_counts['RPM'] <- 10^6*(RocA3_all_counts/RocA3_factor)
DMSO_1_first75['RPM'] <- 10^6*(DMSO_1_first75/DMSO_1_factor)
DMSO_2_first75['RPM'] <- 10^6*(DMSO_2_first75/DMSO_2_factor)
RocA003_first75['RPM'] <- 10^6*(RocA003_first75/RocA003_factor)
RocA03_first75['RPM'] <- 10^6*(RocA03_first75/RocA03_factor)
RocA3_first75['RPM'] <- 10^6*(RocA3_first75/RocA3_factor)
## mean value of DMSO
DMSO_all_counts <- (DMSO_1_all_counts+DMSO_2_all_counts)/2
DMSO_first75 <- (DMSO_1_first75+DMSO_2_first75)/2
## construct dataframe
all_RPM <- data.frame(DMSO=DMSO_all_counts$RPM,RocA003=RocA003_all_counts$RPM,RocA03=RocA03_all_counts$RPM,RocA3=RocA3_all_counts$RPM)
rownames(all_RPM) <- rownames(RocA003_all_counts)
first75_RPM <- data.frame(DMSO=DMSO_first75$RPM,RocA003=RocA003_first75$RPM,RocA03=RocA03_first75$RPM,RocA3=RocA3_first75$RPM)
rownames(first75_RPM) <- rownames(RocA003_all_counts)
after75_RPM <- all_RPM-first75_RPM
## select common transcript used for analysis, all_CDS_unnormed_transcript_id.txt generated by Figure 1B results.
filtered_trans <- read.table("all_CDS_unnormed_transcript_id.txt",sep="\t",stringsAsFactors = F,header = T)
common_trans <- Reduce(intersect,filtered_trans)
all_RPM_common <- all_RPM[common_trans,]
first75_RPM_common <- first75_RPM[common_trans,]
after75_RPM_common <- after75_RPM[common_trans,]
## visulization
library(ggpubr)
library(reshape2)
library(export)
ratio_reshaped <- melt(ratio)
p<-ggboxplot(ratio_reshaped,x="variable",y="value",fill="variable",palette = "npg",xlab = FALSE,ylab = "Counts ratio of the first 75 codons",size=1,font.label = 16)
pp <- ggpar(p,legend = "right",legend.title = "Samples")
pp
graph2ppt(x=pp,"../results/Figure 1C.ppt")
- DE analysis via DEseq2.R
setwd("../data")
## read data
RNA <- read.table("RNA.txt",sep="\t",stringsAsFactors = F,header = T,check.names = F)
rownames(RNA) <- RNA$gene_id
RNA <- RNA[,-1]
RPF <- read.table("RPF.txt",sep="\t",stringsAsFactors = F,header = T,check.names = F)
rownames(RPF) <- RPF$gene_id
RPF <- RPF[,-1]
human_info <- read.table("Homo_sapiens.GRCh38.88.transInfo.txt",sep="\t",stringsAsFactors = F,check.names = F,header = T)
longest_info <- read.table("longest.transcripts.info.txt",sep="\t",stringsAsFactors = F,check.names = F,header = T)
common_gene_id <- intersect(rownames(RPF),rownames(RNA))
RNA_common <- RNA[common_gene_id,]
RPF_common <- RPF[common_gene_id,]
common_gene_info <- human_info[which(human_info$gene_id%in%common_gene_id),]
## DE
library(DESeq2)
RocA03_RPF <- RPF[,c(1,2,4)]
indice <- Reduce(intersect,lapply(RocA03_RPF,function(x){which(x>=10)}))
RPF_filtered <- RocA03_RPF[indice,]
RPF_filtered_DESeq2 <- RPF_filtered
# construct sample information
colData <- data.frame(sample=colnames(RPF_filtered_DESeq2),condition=rep(c("DMSO","RocA03"),times=c(2,1)))
rownames(colData) <- as.character(colData$sample)
# construct DESeq dataset
dds <- DESeqDataSetFromMatrix(countData = RPF_filtered_DESeq2,colData = colData,design = ~condition)
# normalization
dds <- DESeq(dds)
# get the results
res <- results(dds)
summary(res)
head(res)
# get differential expression genes
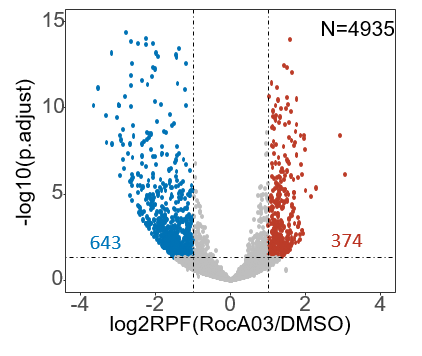
table(res$padj<0.05)
res <- res[order(res$padj),]
DE_genes_table <- subset(res,padj < 0.05 & abs(log2FoldChange)>=1)
write.table(DE_genes_table,"DE_genes_table.txt",sep="\t",quote = F,row.names = TRUE,col.names = TRUE)
DE_genes <- rownames(DE_genes_table)
# get expression matrix of DE genes(normalized) and counts
DE_genes_counts_normalized <- counts(dds,normalize=TRUE)[DE_genes,]
DE_genes_counts <- RPF_filtered_DESeq2[DE_genes,]
up_genes_res <- as.data.frame(subset(res,padj<=0.05&log2FoldChange>=1))
down_genes_res <- as.data.frame(subset(res,padj<=0.05&log2FoldChange<=-1))
Unchanged_genes_res <- as.data.frame(res)[setdiff(res@rownames,union(rownames(up_genes_res),rownames(down_genes_res))),]
up_gene_info <- longest_info[match(rownames(up_genes_res),longest_info$gene_id),]
down_gene_info <- longest_info[match(rownames(down_genes_res),longest_info$gene_id),]
unchanged_gene_info <- longest_info[match(rownames(Unchanged_genes_res),longest_info$gene_id),]
write.table(up_gene_info,"374_up_genes_info.txt",sep="\t",quote=F,row.names = F,col.names = T)
write.table(down_gene_info,"643_down_genes_info.txt",sep="\t",quote=F,row.names = F,col.names = T)
write.table(unchanged_gene_info,"3918_unchanged_genes_info.txt",sep="\t",quote=F,row.names = F,col.names = T)
write.table(up_genes_res,"374_up_genes_data.txt",sep="\t",quote=F,row.names = T,col.names = T)
write.table(down_genes_res,"643_down_genes_data.txt",sep="\t",quote=F,row.names = T,col.names = T)
- visualization
## volcano plot using ggplot2
log2FC <- res$log2FoldChange
padjust <- res$padj
data_for_plot <- data.frame(log2FC=log2FC,padjust=padjust)
data_for_plot <- na.omit(data_for_plot)
data_for_plot$sig[(data_for_plot$padjust>0.05|data_for_plot$padjust=="NA")|(data_for_plot$log2FC>-1|data_for_plot$log2FC<1)] <- "NO"
data_for_plot$sig[(data_for_plot$padjust<=0.05&data_for_plot$log2FC>=1)] <- "up"
data_for_plot$sig[(data_for_plot$padjust<=0.05 & data_for_plot$log2FC<=-1)] <- "down"
summary(data_for_plot$sig=="up")
summary(data_for_plot$sig=="down")
summary(data_for_plot$sig=="NO")
x_lim <- max(log2FC,-log2FC)
library(ggplot2)
library(RColorBrewer)
theme_set(theme_bw())
p <- ggplot(data_for_plot,aes(log2FC,-1*log10(padjust),color = sig))+geom_point()+xlim(-4,4)+labs(x="log2RPF(RocA03/DMSO)",y="-log10(p.adjust)")
p <- p + scale_color_manual(values =c("#0072B5","grey","#BC3C28"))+
geom_hline(yintercept=-log10(0.05),linetype=4)+
geom_vline(xintercept=c(-1,1),linetype=4)
p <- p +theme(panel.grid =element_blank())+
theme(axis.line = element_line(size=0))+ylim(0,15)
p <- p +guides(colour = FALSE)
p <- p +theme(axis.text=element_text(size=20),axis.title=element_text(size=20))
p
- Metagene analysis via RiboMiner
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA
attribute=$workdir/GSE70211_configure.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='DMSO,RocA003,RocA03,RocA3'
replicates='DMSO-1,DMSO-2__RocA-003__RocA-03__RocA-3'
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_RocA03_up_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 0 -e 5 --norm no -y 100 --CI 0.95 --type UTR -S 374_up_genes_info.txt
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_RocA03_down_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 0 -e 5 --norm no -y 100 --CI 0.95 --type UTR -S 643_down_genes_info.txt
PlotMetageneAnalysis -i $results/MA_RocA03_up_unnormed_dataframe.txt -o $results/MA_RocA03_up_unnormed -g $groups -r $replicates -u 100 -d 400 --mode mean
PlotMetageneAnalysis -i $results/MA_RocA03_down_unnormed_dataframe.txt -o $results/MA_RocA03_down_unnormed -g $groups -r $replicates -u 100 -d 400 --mode mean
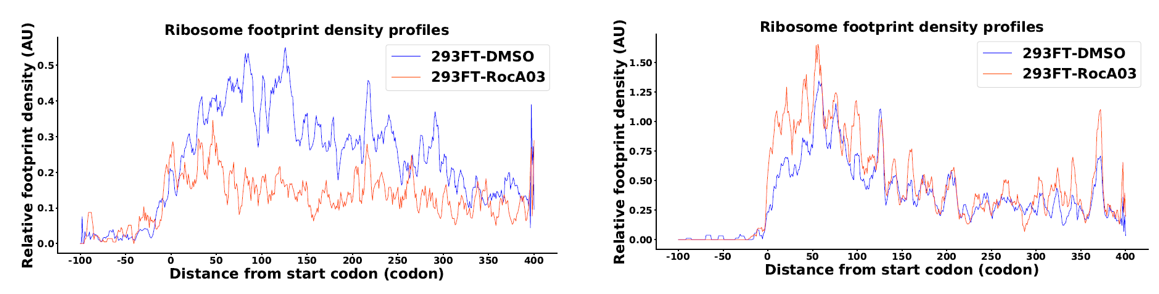
Disome-seq data analysis was finished by previous snakemake pipeline.
bamFiles readLengths Offsets bamLegends
../07.STAR/293FT-DMSO-disome-1_STAR/293FT-DMSO-disome-1.Aligned.toTranscriptome.out.sorted.bam ALL 0 293FT-DMSO-1
../07.STAR/293FT-RocA03-disome-1_STAR/293FT-RocA03-disome-1.Aligned.toTranscriptome.out.sorted.bam ALL 0 293FT-RocA03-1
../07.STAR/293FT-RocA03-disome-2_STAR/293FT-RocA03-disome-2.Aligned.toTranscriptome.out.sorted.bam ALL 0 293FT-RocA03-2
- Metagene plot via RiboMiner
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA
attribute=$workdir/Disome_configure.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='293FT-DMSO,293FT-RocA03'
replicates='293FT-DMSO-1__293FT-RocA03-1,293FT-RocA03-2'
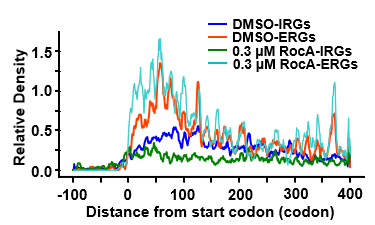
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_RocA03_RUG_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 1 -e 30 --norm no -y 100 --CI 0.95 --type UTR -S 374_up_genes_info.txt
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_RocA03_RDG_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 1 -e 30 --norm no -y 100 --CI 0.95 --type UTR -S 643_down_genes_info.txt
PlotMetageneAnalysis -i $results/MA_RocA03_RUG_unnormed_dataframe.txt -o $results/MA_RocA03_RUG_unnormed -g $groups -r $replicates -u 100 -d 400 --mode mean --slide-window y
PlotMetageneAnalysis -i $results/MA_RocA03_RDG_unnormed_dataframe.txt -o $results/MA_RocA03_RDG_unnormed -g $groups -r $replicates -u 100 -d 400 --mode mean --slide-window y
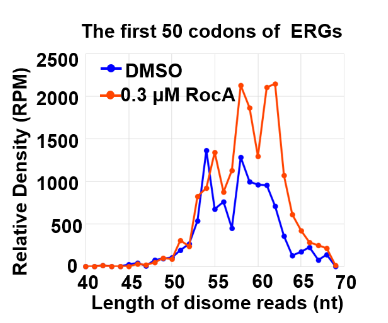
- Calculate disome density of the first 50 codons via ReadsLengthOfSpecificRegions.py
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA_3D
attribute=$workdir/Disome_configure.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='293FT-DMSO,293FT-RocA03'
replicates='293FT-DMSO-1__293FT-RocA03-1,293FT-RocA03-2'
up_trans=374_up_genes_info.txt
down_trans=643_down_genes_info.txt
mkdir $workdir/Length
python ReadsLengthOfSpecificRegions.py -i $BamDir/293FT-DMSO-disome-1_STAR/293FT-DMSO-disome-1.Aligned.toTranscriptome.out.sorted.bam -o $workdir/Length/293FT_DMSO_1_RUG_100codons -c $trans_info --type CDS -S 374_up_genes_info.txt -l 1 -r 150
python ReadsLengthOfSpecificRegions.py -i $BamDir/293FT-RocA03-disome-1_STAR/293FT-RocA03-disome-1.Aligned.toTranscriptome.out.sorted.bam -o $workdir/Length/293FT_RocA03_1_RUG_100codons -c $trans_info --type CDS -S 374_up_genes_info.txt -l 1 -r 150
python ReadsLengthOfSpecificRegions.py -i $BamDir/293FT-RocA03-disome-2_STAR/293FT-RocA03-disome-2.Aligned.toTranscriptome.out.sorted.bam -o $workdir/Length/293FT_RocA03_2_RUG_100codons -c $trans_info --type CDS -S 374_up_genes_info.txt -l 1 -r 150
## plot in MicroSoft Excell
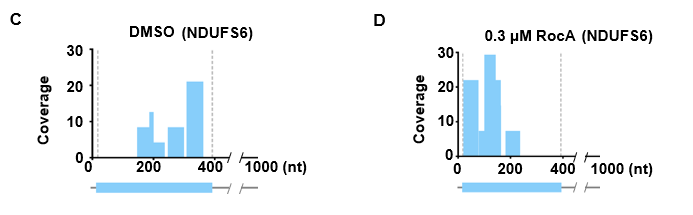
- Calculate read coverage for each transcript via RiboMiner
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA
attribute=$workdir/Disome_configure.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='293FT-DMSO,293FT-RocA03,HT-29-DMSO,HT-29-RocA03'
replicates='293FT-DMSO__293FT-RocA03__HT-29-DMSO__HT-29-RocA03'
CoverageOfEachTrans -f $attribute -c $trans_info -o coverage/MA_RUG --mode coverage -S 374_up_genes_info.txt
PlotTransCoverage -i coverage/MA_RUG_293FT-DMSO_RPM_depth.txt -o coverage/293FT_DMSO_NDUFS6 -c $trans_info -t NDUFS6 --mode coverage --id-type gene_name --color lightskyblue --type single-gene --ymax 30
PlotTransCoverage -i coverage/MA_RUG_293FT-RocA03_RPM_depth.txt -o coverage/293FT_RocA03_NDUFS6 -c $trans_info -t NDUFS6 --mode coverage --id-type gene_name --color lightskyblue --type single-gene --ymax 30
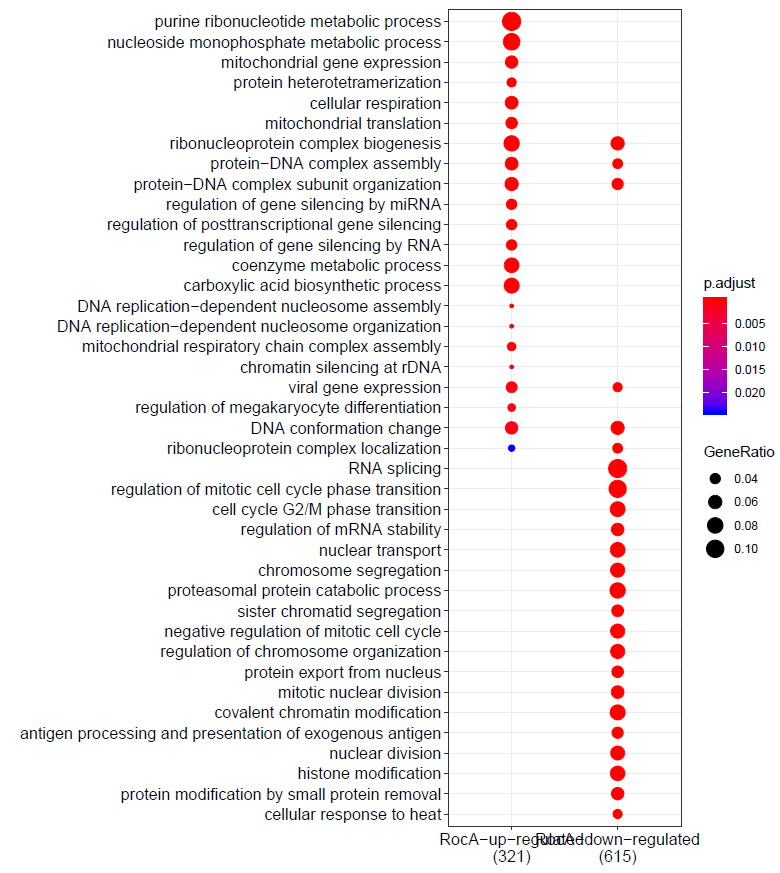
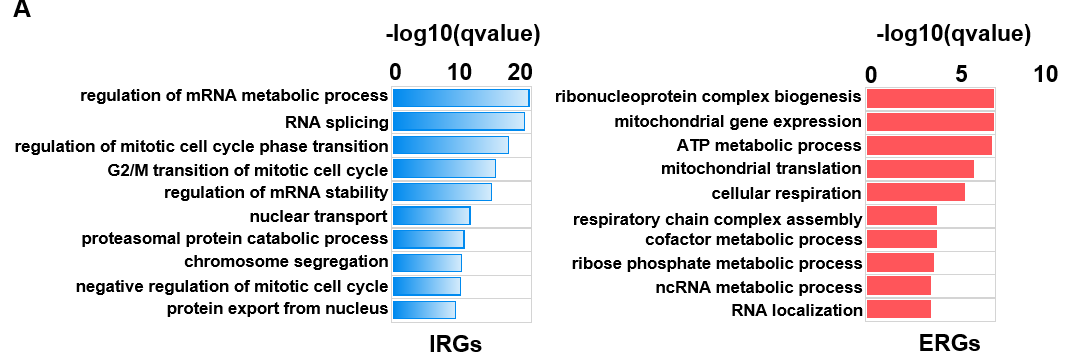
- Get gene sets for GO analysis: GO_analysis.txt
$ less -S GO_analysis.txt
RocA-up-regulated RocA-down-regulated
MT-ND5 RBBP7
MT-ATP6 KPNB1
MT-CYB HNRNPD
MT-ND4 HNRNPAB
MT-ND1 SLC1A5
HIST1H3B HUWE1
MT-CO3 XPO1
MT-ND2 HNRNPF
C1QBP ATP2A2
MT-CO2 CLTC
GPI ZNF711
HIST1H4B H2AFY
LDHA NUCKS1
DYNC1H1 MYC
HIST1H2AH CTNNB1
NUP205 YWHAE
- Codes: GO_analysis.R
setwd("../data")
if (!require("BiocManager", quietly = TRUE))
install.packages("BiocManager")
BiocManager::install("clusterProfiler")
library(clusterProfiler)
library(org.Hs.eg.db)
GO_analysis <- read.table("GO_analysis.txt",sep="\t",stringsAsFactors = F,header = T,fill = T,check.names = F)
RocA_insensitive <- na.omit(GO_analysis$`RocA-up-regulated`)
RocA_sensitive <- na.omit(GO_analysis$`RocA-down-regulated`)
## construct dataframe
# up
RocA_insensitive_entrezID <- bitr(RocA_insensitive,fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")
group_up <- rep("RocA-up-regulated",times=length(RocA_insensitive_entrezID$ENTREZID))
data_up <- data.frame(geneID=RocA_insensitive_entrezID$ENTREZID,group=group_up)
# down
RocA_sensitive_entrezID <- bitr(RocA_sensitive,fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Hs.eg.db")
group_down <- rep("RocA-down-regulated",times=length(RocA_sensitive_entrezID$ENTREZID))
data_down <- data.frame(geneID=RocA_sensitive_entrezID$ENTREZID,group=group_down)
# all
data_all_GO <- rbind(data_up,data_down)
formula <- compareCluster(geneID~group, data=data_all_GO, fun='enrichGO',OrgDb='org.Hs.eg.db',pvalueCutoff=0.05,pAdjustMethod = "BH",qvalueCutoff = 0.05,ont="BP")
summary(formula)
formula <- simplify(formula)
y <- dotplot(formula,showCategory=20,includeAll=TRUE)
y
formula_dataframe <- as.data.frame(formula)
write.table(formula_dataframe,"GO_analysis_dataframe.txt",quote = F,sep="\t",row.names = F,col.names = T)
regenated in Excel:
- Fetch cds sequences for RUGs and RDGs via RiboMiner
$ GetProteinCodingSequence -i ./data/RiboCode_annot/transcripts_sequence.fa -c ./data/longest.transcripts.info.txt -o 643_down --mode whole --table 1 -S 643_down_genes_info.txt
19636 transcripts will be used in the follow analysis.
There are 642 transcripts from 643_down_genes_info.txt used for following analysis.
ENST00000625083 filtered--There is a ambiguous nucleotide N in your sequence
ENST00000387347 filtered--There is a ambiguous nucleotide N in your sequence
Warning: sequence is not divisible by 3
Warning: sequence is not divisible by 3
Warning: sequence is not divisible by 3
Notes: There are 3 transcripts whose cds sequence cannot be divided by 3!
Finish the step of extracting sequences!
$ GetProteinCodingSequence -i ./data/RiboCode_annot/transcripts_sequence.fa -c ./data/longest.transcripts.info.txt -o 374_up --mode whole --table 1 -S 374_up_genes_info.txt 19636 transcripts will be used in the follow analysis.
There are 374 transcripts from 374_up_genes_info.txt used for following analysis.
ENST00000625083 filtered--There is a ambiguous nucleotide N in your sequence
ENST00000387347 filtered--There is a ambiguous nucleotide N in your sequence
Warning: sequence is not divisible by 3
Warning: sequence is not divisible by 3
Warning: sequence is not divisible by 3
Warning: sequence is not divisible by 3
Warning: sequence is not divisible by 3
Warning: sequence is not divisible by 3
Notes: There are 6 transcripts whose cds sequence cannot be divided by 3!
Finish the step of extracting sequences!
$ GetProteinCodingSequence -i ./data/RiboCode_annot/transcripts_sequence.fa -c ./data/longest.transcripts.info.txt -o MTGenes --mode whole --table 1 -S MTGene_info.txt
- Calculate tAI, CAI, and charges
workdir=.
fasta=./data
Ref=/workdata/LabMember2/lifj/lifj/data/Reference/human
## 1 cAI
cAI -i MT_Genes_cds_sequences.fa,$fasta/374_up_cds_sequences.fa,$fasta/643_down_cds_sequences.fa,$Ref/longest_cds_sequences.fa -o CAI -t MTGenes,RocA-up-regulated,RocA-down-regulated,all -u 0 -d 100 --reference $Ref/longest_cds_sequences.fa
cAIPlot -i CAI_local_cAI_dataframe.txt -o CAI -u 0 -d 100 --mode all --start 5 --window 7 --step 1
## 2 tAI
tAI -i MT_Genes_cds_sequences.fa,$fasta/374_up_cds_sequences.fa,$fasta/643_down_cds_sequences.fa,$Ref/longest_cds_sequences.fa -o tAI -t MTGenes,RocA-up-regulated,RocA-down-regulated,all -u 0 -d 100 --table 1 -N tRNA_GCNs_human.txt
tAIPlot -i tAI_tAI_dataframe.txt -o tAI -u 0 -d 00 --mode all --start 5 --window 7 --step 1
## 3 hydropathy and charge
hydropathyCharge -i MT_Genes_cds_sequences.fa,$fasta/374_up_cds_sequences.fa,$fasta/643_down_cds_sequences.fa,$Ref/longest_cds_sequences.fa -o Hydro -t MTGenes,RocA-up-regulated,RocA-down-regulated,all --index hydropathy.txt -u 0 -d 100 --table 1
hydropathyCharge -i MT_Genes_cds_sequences.fa,$fasta/374_up_cds_sequences.fa,$fasta/643_down_cds_sequences.fa,$Ref/longest_cds_sequences.fa -o Charge -t MTGenes,RocA-up-regulated,RocA-down-regulated,all --index AA_charge.txt -u 0 -d 100 --table 1
PlotHydropathyCharge -i Hydro_values_dataframe.txt -o Hydro -u 0 -d 100 --mode all --ylab "Average Hydrophobicity"
PlotHydropathyCharge -i Charge_values_dataframe.txt -o Charge -u 0 -d 100 --mode all --ylab "Average Charges"
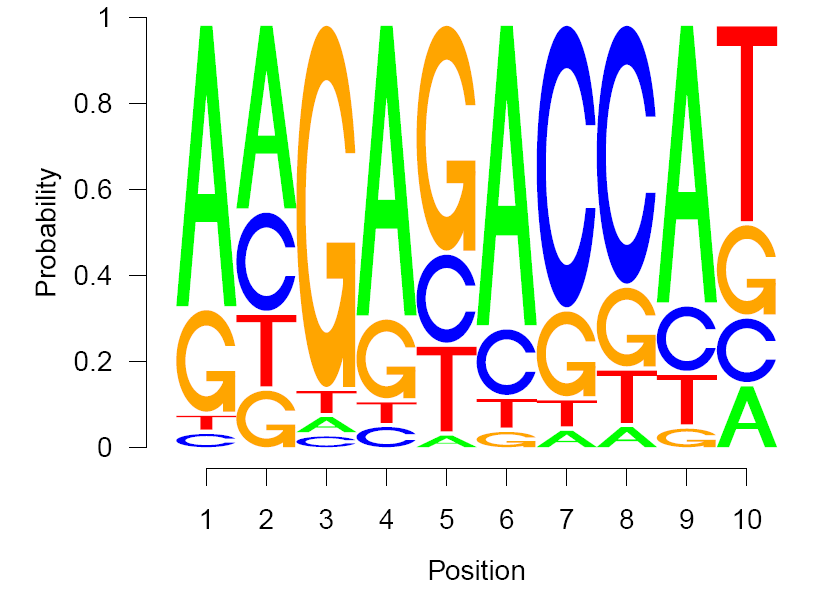
- Motif identification via homer
$ cat run_homer.sh
findMotifs.pl 374_up_CDS.fa human up_CDS -fasta ~/Reference/human/longest_CDS.fa
findMotifs.pl 643_down_CDS.fa human down_CDS -fasta ~/Reference/human/longest_CDS.fa
$ cat homer-RocA03-up-CDS-motif.txt
A C G T
0.686 0.032 0.248 0.034
0.447 0.240 0.138 0.175
0.037 0.026 0.884 0.053
0.709 0.049 0.192 0.050
0.028 0.215 0.550 0.207
0.733 0.160 0.037 0.070
0.040 0.689 0.207 0.064
0.049 0.631 0.192 0.128
0.677 0.157 0.045 0.121
0.149 0.155 0.218 0.477
- Select one polypurine motif in CDS of RUGs for plot via Seq2logo.R
setwd("../data")
library(seqLogo)
up_motif3 <- read.table("homer-RocA03-up-CDS-motif.txt",sep="\t",stringsAsFactors = F,header = T)
up_motif3 <- t(up_motif3)
seqLogo(up_motif3,ic.scale = F)
- blast iCLIP-seq reads to CDS of RUGs
bsub -q TEST-A -n 4 -e 374_up.err -o 374_up.out "blastn -subject 374_up_CDS.fa -query ../05.contam/noncontam_SRR3238818.fa -task blastn -outfmt \"7 qseqid sseqid pident length mismatch gapopen qstart qend sstart send evalue\" -out RocA3_to_RocA03_up_CDS.txt"
bsub -q TEST-A -n 4 -e 374_up.err -o 374_up.out "blastn -subject 374_up_CDS.fa -query ../05.contam/noncontam_SRR3238817.fa -task blastn -outfmt \"7 qseqid sseqid pident length mismatch gapopen qstart qend sstart send evalue\" -out RocA03_to_RocA03_up_CDS.txt"
bsub -q TEST-A -n 4 -e 374_up.err -o 374_up.out "blastn -subject 374_up_CDS.fa -query ../05.contam/noncontam_SRR3238816.fa -task blastn -outfmt \"7 qseqid sseqid pident length mismatch gapopen qstart qend sstart send evalue\" -out RocA003_to_RocA03_up_CDS.txt"
bsub -q TEST-A -n 4 -e 374_up.err -o 374_up.out "blastn -subject 374_up_CDS.fa -query ../05.contam/noncontam_SRR3238815.fa -task blastn -outfmt \"7 qseqid sseqid pident length mismatch gapopen qstart qend sstart send evalue\" -out DMSO2_to_RocA03_up_CDS.txt"
bsub -q TEST-A -n 4 -e 374_up.err -o 374_up.out "blastn -subject 374_up_CDS.fa -query ../05.contam/noncontam_SRR3238814.fa -task blastn -outfmt \"7 qseqid sseqid pident length mismatch gapopen qstart qend sstart send evalue\" -out DMSO1_to_RocA03_up_CDS.txt"
- blast iCLIP-seq reads to 5UTR of RDGs
bsub -q TEST-A -n 4 -e 643_down.err -o 643_down.out "blastn -subject 643_down_5UTR.fa -query ../05.contam/noncontam_SRR3238818.fa -task blastn -outfmt \"7 qseqid sseqid pident length mismatch gapopen qstart qend sstart send evalue\" -out RocA3_to_RocA03_down_5UTR.txt"
bsub -q TEST-A -n 4 -e 643_down.err -o 643_down.out "blastn -subject 643_down_5UTR.fa -query ../05.contam/noncontam_SRR3238817.fa -task blastn -outfmt \"7 qseqid sseqid pident length mismatch gapopen qstart qend sstart send evalue\" -out RocA03_to_RocA03_down_5UTR.txt"
bsub -q TEST-A -n 4 -e 643_down.err -o 643_down.out "blastn -subject 643_down_5UTR.fa -query ../05.contam/noncontam_SRR3238816.fa -task blastn -outfmt \"7 qseqid sseqid pident length mismatch gapopen qstart qend sstart send evalue\" -out RocA003_to_RocA03_down_5UTR.txt"
bsub -q TEST-A -n 4 -e 643_down.err -o 643_down.out "blastn -subject 643_down_5UTR.fa -query ../05.contam/noncontam_SRR3238815.fa -task blastn -outfmt \"7 qseqid sseqid pident length mismatch gapopen qstart qend sstart send evalue\" -out DMSO2_to_RocA03_down_5UTR.txt"
bsub -q TEST-A -n 4 -e 643_down.err -o 643_down.out "blastn -subject 643_down_5UTR.fa -query ../05.contam/noncontam_SRR3238814.fa -task blastn -outfmt \"7 qseqid sseqid pident length mismatch gapopen qstart qend sstart send evalue\" -out DMSO1_to_RocA03_down_5UTR.txt"
- Searching polyAG motifs via SearchPolypurineMotifs.py
python SearchPolypurineMotifs.py -i DMSO1_to_RocA03_up_CDS_reads.fa -o DMSO1_to_RocA03_up_CDS_reads --kmer 4 --base AG
python SearchPolypurineMotifs.py -i DMSO2_to_RocA03_up_CDS_reads.fa -o DMSO2_to_RocA03_up_CDS_reads --kmer 4 --base AG
python SearchPolypurineMotifs.py -i RocA003_to_RocA03_up_CDS_reads.fa -o RocA003_to_RocA03_up_CDS_reads --kmer 4 --base AG
python SearchPolypurineMotifs.py -i RocA03_to_RocA03_up_CDS_reads.fa -o RocA03_to_RocA03_up_CDS_reads --kmer 4 --base AG
python SearchPolypurineMotifs.py -i RocA3_to_RocA03_up_CDS_reads.fa -o RocA3_to_RocA03_up_CDS_reads --kmer 4 --base AG
python SearchPolypurineMotifs.py -i DMSO1_to_RocA03_down_5UTR_reads.fa -o DMSO1_to_RocA03_down_5UTR_reads --kmer 4 --base AG
python SearchPolypurineMotifs.py -i DMSO2_to_RocA03_down_5UTR_reads.fa -o DMSO2_to_RocA03_down_5UTR_reads --kmer 4 --base AG
python SearchPolypurineMotifs.py -i RocA003_to_RocA03_down_5UTR_reads.fa -o RocA003_to_RocA03_down_5UTR_reads --kmer 4 --base AG
python SearchPolypurineMotifs.py -i RocA03_to_RocA03_down_5UTR_reads.fa -o RocA03_to_RocA03_down_5UTR_reads --kmer 4 --base AG
python SearchPolypurineMotifs.py -i RocA3_to_RocA03_down_5UTR_reads.fa -o RocA3_to_RocA03_down_5UTR_reads --kmer 4 --base AG
python SearchPolypurineMotifs.py -i DMSO1_to_RocA03_up_CDS_reads.fa -o DMSO1_to_RocA03_up_CDS_reads --kmer 6 --base AG
python SearchPolypurineMotifs.py -i DMSO2_to_RocA03_up_CDS_reads.fa -o DMSO2_to_RocA03_up_CDS_reads --kmer 6 --base AG
python SearchPolypurineMotifs.py -i RocA003_to_RocA03_up_CDS_reads.fa -o RocA003_to_RocA03_up_CDS_reads --kmer 6 --base AG
python SearchPolypurineMotifs.py -i RocA03_to_RocA03_up_CDS_reads.fa -o RocA03_to_RocA03_up_CDS_reads --kmer 6 --base AG
python SearchPolypurineMotifs.py -i RocA3_to_RocA03_up_CDS_reads.fa -o RocA3_to_RocA03_up_CDS_reads --kmer 6 --base AG
python SearchPolypurineMotifs.py -i DMSO1_to_RocA03_up_CDS_reads.fa -o DMSO1_to_RocA03_up_CDS_reads --kmer 5 --base AG
python SearchPolypurineMotifs.py -i DMSO2_to_RocA03_up_CDS_reads.fa -o DMSO2_to_RocA03_up_CDS_reads --kmer 5 --base AG
python SearchPolypurineMotifs.py -i RocA003_to_RocA03_up_CDS_reads.fa -o RocA003_to_RocA03_up_CDS_reads --kmer 5 --base AG
python SearchPolypurineMotifs.py -i RocA03_to_RocA03_up_CDS_reads.fa -o RocA03_to_RocA03_up_CDS_reads --kmer 5 --base AG
python SearchPolypurineMotifs.py -i RocA3_to_RocA03_up_CDS_reads.fa -o RocA3_to_RocA03_up_CDS_reads --kmer 5 --base AG
python SearchPolypurineMotifs.py -i DMSO1_to_RocA03_down_5UTR_reads.fa -o DMSO1_to_RocA03_down_5UTR_reads --kmer 6 --base AG
python SearchPolypurineMotifs.py -i DMSO2_to_RocA03_down_5UTR_reads.fa -o DMSO2_to_RocA03_down_5UTR_reads --kmer 6 --base AG
python SearchPolypurineMotifs.py -i RocA003_to_RocA03_down_5UTR_reads.fa -o RocA003_to_RocA03_down_5UTR_reads --kmer 6 --base AG
python SearchPolypurineMotifs.py -i RocA03_to_RocA03_down_5UTR_reads.fa -o RocA03_to_RocA03_down_5UTR_reads --kmer 6 --base AG
python SearchPolypurineMotifs.py -i RocA3_to_RocA03_down_5UTR_reads.fa -o RocA3_to_RocA03_down_5UTR_reads --kmer 6 --base AG
python SearchPolypurineMotifs.py -i DMSO1_to_RocA03_down_5UTR_reads.fa -o DMSO1_to_RocA03_down_5UTR_reads --kmer 5 --base AG
python SearchPolypurineMotifs.py -i DMSO2_to_RocA03_down_5UTR_reads.fa -o DMSO2_to_RocA03_down_5UTR_reads --kmer 5 --base AG
python SearchPolypurineMotifs.py -i RocA003_to_RocA03_down_5UTR_reads.fa -o RocA003_to_RocA03_down_5UTR_reads --kmer 5 --base AG
python SearchPolypurineMotifs.py -i RocA03_to_RocA03_down_5UTR_reads.fa -o RocA03_to_RocA03_down_5UTR_reads --kmer 5 --base AG
python SearchPolypurineMotifs.py -i RocA3_to_RocA03_down_5UTR_reads.fa -o RocA3_to_RocA03_down_5UTR_reads --kmer 5 --base AG
- statistic the frequency of poly-purine motifs
## collapse UTR
less -S RocA3_to_RocA03_down_5UTR_reads_polyAG_4_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA3_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed.txt
less -S RocA3_to_RocA03_down_5UTR_reads_polyAG_5_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA3_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed.txt
less -S RocA3_to_RocA03_down_5UTR_reads_polyAG_6_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA3_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed.txt
less -S RocA03_to_RocA03_down_5UTR_reads_polyAG_4_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA03_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed.txt
less -S RocA03_to_RocA03_down_5UTR_reads_polyAG_5_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA03_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed.txt
less -S RocA03_to_RocA03_down_5UTR_reads_polyAG_6_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA03_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed.txt
less -S RocA003_to_RocA03_down_5UTR_reads_polyAG_4_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA003_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed.txt
less -S RocA003_to_RocA03_down_5UTR_reads_polyAG_5_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA003_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed.txt
less -S RocA003_to_RocA03_down_5UTR_reads_polyAG_6_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA003_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed.txt
less -S DMSO1_to_RocA03_down_5UTR_reads_polyAG_4_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > DMSO1_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed.txt
less -S DMSO1_to_RocA03_down_5UTR_reads_polyAG_5_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > DMSO1_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed.txt
less -S DMSO1_to_RocA03_down_5UTR_reads_polyAG_6_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > DMSO1_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed.txt
less -S DMSO2_to_RocA03_down_5UTR_reads_polyAG_4_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > DMSO2_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed.txt
less -S DMSO2_to_RocA03_down_5UTR_reads_polyAG_5_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > DMSO2_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed.txt
less -S DMSO2_to_RocA03_down_5UTR_reads_polyAG_6_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > DMSO2_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed.txt
## collapse CDS
less -S RocA3_to_RocA03_up_CDS_reads_polyAG_4_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA3_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed.txt
less -S RocA3_to_RocA03_up_CDS_reads_polyAG_5_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA3_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed.txt
less -S RocA3_to_RocA03_up_CDS_reads_polyAG_6_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA3_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed.txt
less -S RocA03_to_RocA03_up_CDS_reads_polyAG_4_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA03_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed.txt
less -S RocA03_to_RocA03_up_CDS_reads_polyAG_5_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA03_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed.txt
less -S RocA03_to_RocA03_up_CDS_reads_polyAG_6_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA03_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed.txt
less -S RocA003_to_RocA03_up_CDS_reads_polyAG_4_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA003_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed.txt
less -S RocA003_to_RocA03_up_CDS_reads_polyAG_5_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA003_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed.txt
less -S RocA003_to_RocA03_up_CDS_reads_polyAG_6_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > RocA003_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed.txt
less -S DMSO1_to_RocA03_up_CDS_reads_polyAG_4_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > DMSO1_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed.txt
less -S DMSO1_to_RocA03_up_CDS_reads_polyAG_5_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > DMSO1_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed.txt
less -S DMSO1_to_RocA03_up_CDS_reads_polyAG_6_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > DMSO1_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed.txt
less -S DMSO2_to_RocA03_up_CDS_reads_polyAG_4_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > DMSO2_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed.txt
less -S DMSO2_to_RocA03_up_CDS_reads_polyAG_5_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > DMSO2_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed.txt
less -S DMSO2_to_RocA03_up_CDS_reads_polyAG_6_mer.txt|cut -f 3|sort |uniq -c |awk -F " " 'BEGIN{OFS="\t"}{print $2,$1}' |sort > DMSO2_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed.txt
- Calculate motif score via PolypurineMotifScore.py
# statistic motif score
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238814.fa -m DMSO1_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed.txt -o DMSO1_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238815.fa -m DMSO2_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed.txt -o DMSO2_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238816.fa -m RocA003_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed.txt -o RocA003_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238817.fa -m RocA03_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed.txt -o RocA03_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238818.fa -m RocA3_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed.txt -o RocA3_to_RocA03_up_CDS_reads_polyAG_4_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238814.fa -m DMSO1_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed.txt -o DMSO1_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238815.fa -m DMSO2_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed.txt -o DMSO2_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238816.fa -m RocA003_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed.txt -o RocA003_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238817.fa -m RocA03_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed.txt -o RocA03_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238818.fa -m RocA3_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed.txt -o RocA3_to_RocA03_down_5UTR_reads_polyAG_4_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238814.fa -m DMSO1_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed.txt -o DMSO1_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238815.fa -m DMSO2_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed.txt -o DMSO2_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238816.fa -m RocA003_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed.txt -o RocA003_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238817.fa -m RocA03_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed.txt -o RocA03_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238818.fa -m RocA3_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed.txt -o RocA3_to_RocA03_up_CDS_reads_polyAG_5_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238814.fa -m DMSO1_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed.txt -o DMSO1_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238815.fa -m DMSO2_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed.txt -o DMSO2_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238816.fa -m RocA003_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed.txt -o RocA003_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238817.fa -m RocA03_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed.txt -o RocA03_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238818.fa -m RocA3_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed.txt -o RocA3_to_RocA03_down_5UTR_reads_polyAG_5_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238814.fa -m DMSO1_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed.txt -o DMSO1_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238815.fa -m DMSO2_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed.txt -o DMSO2_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238816.fa -m RocA003_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed.txt -o RocA003_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238817.fa -m RocA03_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed.txt -o RocA03_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238818.fa -m RocA3_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed.txt -o RocA3_to_RocA03_up_CDS_reads_polyAG_6_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238814.fa -m DMSO1_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed.txt -o DMSO1_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238815.fa -m DMSO2_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed.txt -o DMSO2_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238816.fa -m RocA003_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed.txt -o RocA003_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238817.fa -m RocA03_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed.txt -o RocA03_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed
python PolypurineMotifScore.py -i ../05.contam/noncontam_SRR3238818.fa -m RocA3_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed.txt -o RocA3_to_RocA03_down_5UTR_reads_polyAG_6_mer_collapsed
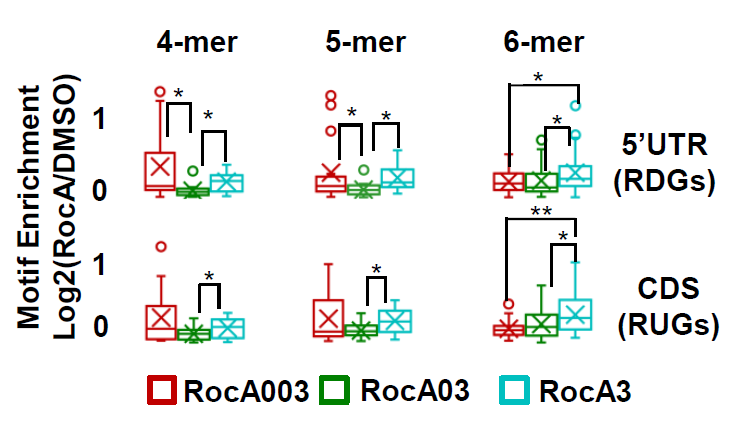
- Calculate motif enrichment (RocA motif score/DMSO motif score) in Excel
./results/Figure 4B.xlsx
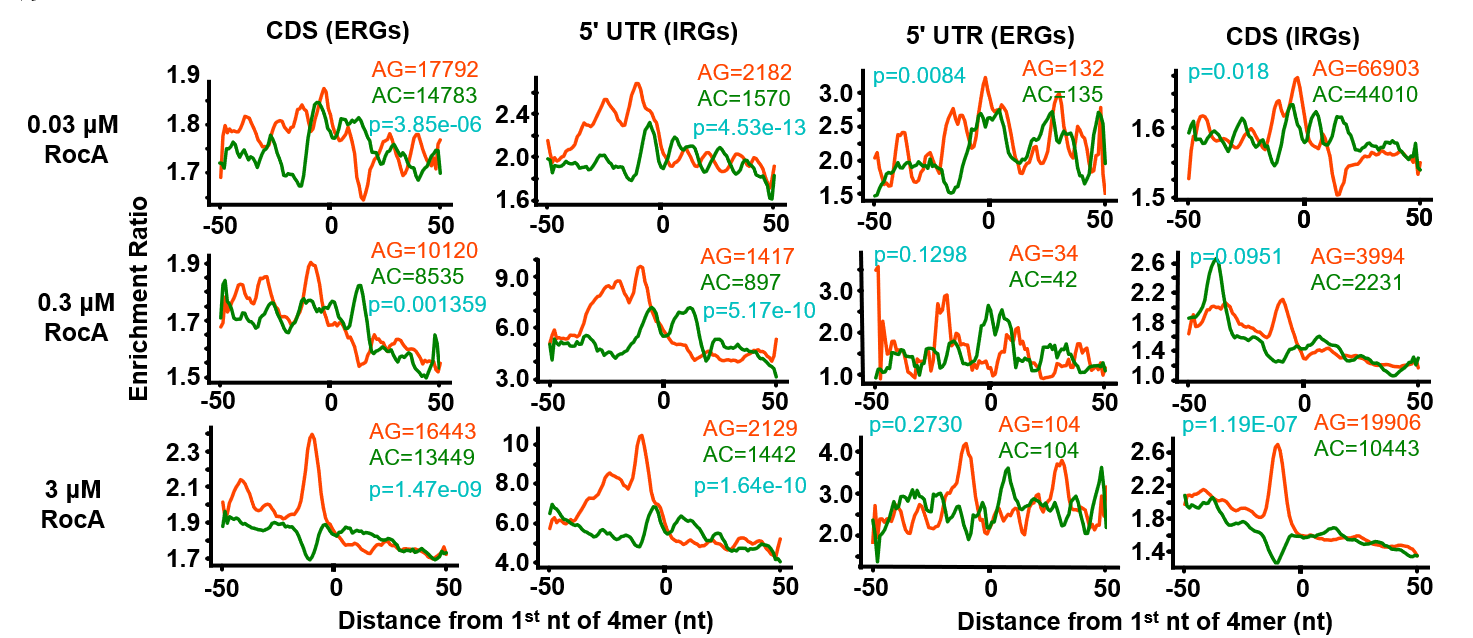
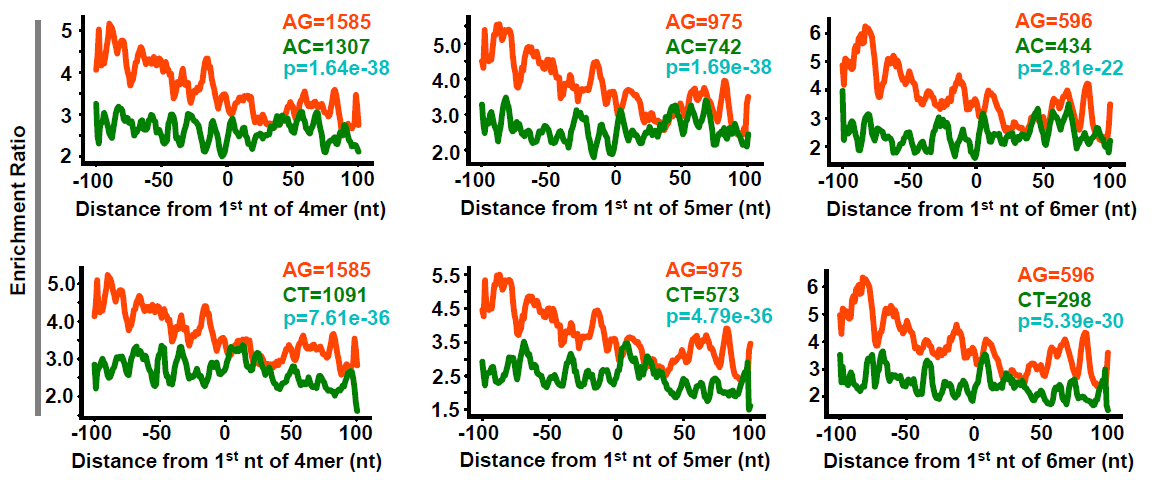
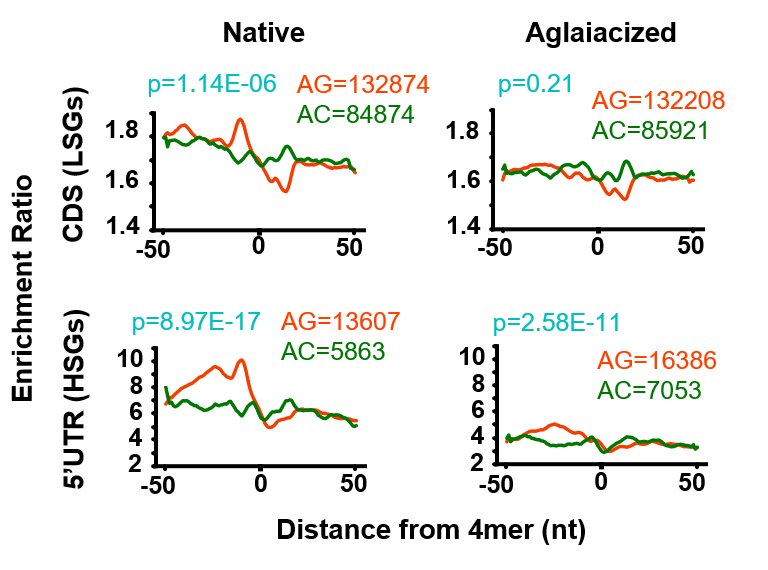
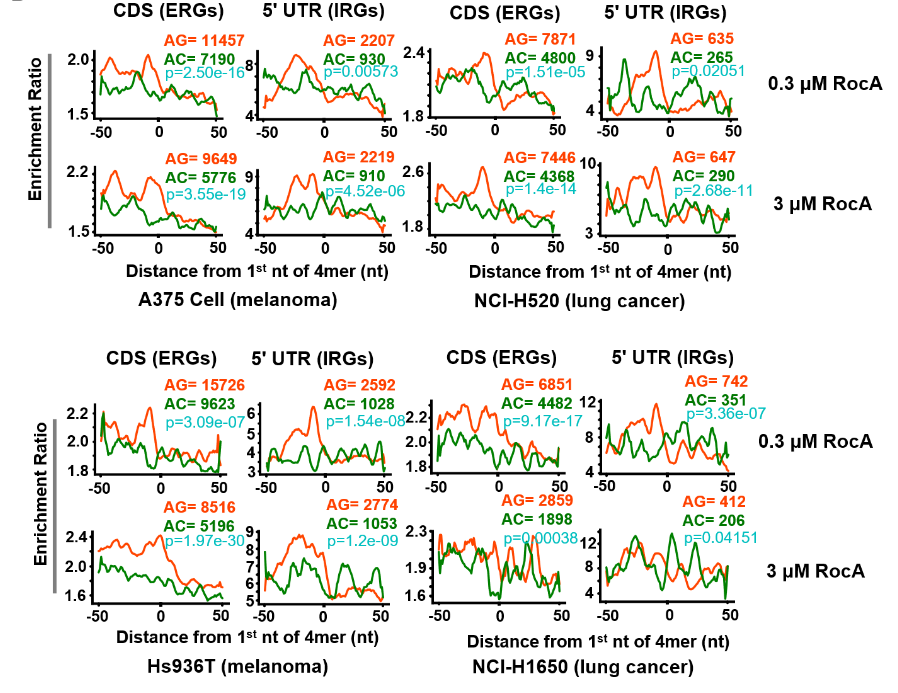
- Calculate density around poly-purine motifs via RiboDensityAroundPolyPurineMotifs.py
workdir=`pwd`
BamDir=/workdata/home/lifj/lifj/Project/05.Ribo_seq_human/GSE70211/07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA_HEK293
attribute=$workdir/GSE70211_configure.txt
trans_info=$Ref/longest.transcripts.info.txt
up_trans=$workdir/374_up_regulated_genes_info.txt
down_trans=$workdir/643_down_regulated_genes_info.txt
## poly-purine
mkdir -p polymotifs/counts_10
## UTR
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t_UTR -M RPKM -u 50 -d 50 -l 0
-n 0 -F $transcript --kmer 4 --type 5UTR -S 643_down_regulated_genes_info.txt --id-type transcript_id --base AG
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t_UTR -M RPKM -u 50 -d 50 -l 0
-n 0 -F $transcript --kmer 4 --type 5UTR -S 643_down_regulated_genes_info.txt --id-type transcript_id --base AC
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t_UTR -M RPKM -u 50 -d 50 -l 0
-n 0 -F $transcript --kmer 4 --type 5UTR -S 643_down_regulated_genes_info.txt --id-type transcript_id --base CT
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t_UTR -M RPKM -u 50 -d 50 -l 0
-n 0 -F $transcript --kmer 5 --type 5UTR -S 643_down_regulated_genes_info.txt --id-type transcript_id --base AG
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t_UTR -M RPKM -u 50 -d 50 -l 0
-n 0 -F $transcript --kmer 5 --type 5UTR -S 643_down_regulated_genes_info.txt --id-type transcript_id --base AC
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t_UTR -M RPKM -u 50 -d 50 -l 0 -n 0 -F $transcript --kmer 5 --type 5UTR -S 643_down_regulated_genes_info.txt --id-type transcript_id --base CT
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t_UTR -M RPKM -u 50 -d 50 -l 0 -n 0 -F $transcript --kmer 6 --type 5UTR -S 643_down_regulated_genes_info.txt --id-type transcript_id --base AG
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t_UTR -M RPKM -u 50 -d 50 -l 0 -n 0 -F $transcript --kmer 6 --type 5UTR -S 643_down_regulated_genes_info.txt --id-type transcript_id --base AC
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t_UTR -M RPKM -u 50 -d 50 -l 0 -n 0 -F $transcript --kmer 6 --type 5UTR -S 643_down_regulated_genes_info.txt --id-type transcript_id --base CT
## CDS
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t -M RPKM -u 50 -d 50 -l 0 -n 0 -F $transcript --kmer 4 --type CDS -S 374_up_regulated_genes_info.txt --id-type transcript_id --base AG
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t -M RPKM -u 50 -d 50 -l 0 -n 0 -F $transcript --kmer 4 --type CDS -S 374_up_regulated_genes_info.txt --id-type transcript_id --base AC
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t -M RPKM -u 50 -d 50 -l 0 -n 0 -F $transcript --kmer 4 --type CDS -S 374_up_regulated_genes_info.txt --id-type transcript_id --base CT
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t -M RPKM -u 50 -d 50 -l 0 -n 0 -F $transcript --kmer 5 --type CDS -S 374_up_regulated_genes_info.txt --id-type transcript_id --base AG
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t -M RPKM -u 50 -d 50 -l 0 -n 0 -F $transcript --kmer 5 --type CDS -S 374_up_regulated_genes_info.txt --id-type transcript_id --base AC
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t -M RPKM -u 50 -d 50 -l 0 -n 0 -F $transcript --kmer 5 --type CDS -S 374_up_regulated_genes_info.txt --id-type transcript_id --base CT
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t -M RPKM -u 50 -d 50 -l 0 -n 0 -F $transcript --kmer 6 --type CDS -S 374_up_regulated_genes_info.txt --id-type transcript_id --base AG
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t -M RPKM -u 50 -d 50 -l 0 -n 0 -F $transcript --kmer 6 --type CDS -S 374_up_regulated_genes_info.txt --id-type transcript_id --base AC
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/hek293t -M RPKM -u 50 -d 50 -l 0 -n 0 -F $transcript --kmer 6 --type CDS -S 374_up_regulated_genes_info.txt --id-type transcript_id --base CT
- polyAG, polyAC and polyCT motifs
$ cat ./data/4mer-5UTR-AG.txt
motif
GGAA
GAAA
AAAA
AGAA
AAGG
AGGA
GAAG
AGAG
AAAG
GAGA
AAGA
AGGG
$ cat ./data/4mer-5UTR-AC.txt
motifs
AAAC
AACA
AACC
ACAA
ACAC
ACCA
ACCC
CAAA
CAAC
CACA
CACC
CCAA
CCAC
CCCA
CCCC
$ cat ./data/4mer-CT.txt
CCCC
CCCT
CCTC
CCTT
CTCC
CTCT
CTTC
CTTT
TCCC
TCCT
TCTC
TCTT
TTCC
TTCT
TTTC
TTTT
- Calculate mean density between replicates
## 01. calculate mean
mkdir counts_10_mean_kmer
## CDS
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_DMSO-1_polyAC_4_mer.txt,counts_10/hek293t_DMSO-2_polyAC_4_mer.txt -o counts_10_mean_kmer/hek293t_DMSO_polyAC_4_mer --kmer 4mer-CDS-AC.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_DMSO-1_polyAG_4_mer.txt,counts_10/hek293t_DMSO-2_polyAG_4_mer.txt -o counts_10_mean_kmer/hek293t_DMSO_polyAG_4_mer --kmer 4mer-CDS-AG.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_DMSO-1_polyCT_4_mer.txt,counts_10/hek293t_DMSO-2_polyCT_4_mer.txt -o counts_10_mean_kmer/hek293t_DMSO_polyCT_4_mer --kmer 4mer-CT.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_DMSO-1_polyAC_5_mer.txt,counts_10/hek293t_DMSO-2_polyAC_5_mer.txt -o counts_10_mean_kmer/hek293t_DMSO_polyAC_5_mer --kmer 5mer-CDS-AC.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_DMSO-1_polyAG_5_mer.txt,counts_10/hek293t_DMSO-2_polyAG_5_mer.txt -o counts_10_mean_kmer/hek293t_DMSO_polyAG_5_mer --kmer 5mer-CDS-AG.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_DMSO-1_polyCT_5_mer.txt,counts_10/hek293t_DMSO-2_polyCT_5_mer.txt -o counts_10_mean_kmer/hek293t_DMSO_polyCT_5_mer --kmer 5mer-CT.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_DMSO-1_polyAC_6_mer.txt,counts_10/hek293t_DMSO-2_polyAC_6_mer.txt -o counts_10_mean_kmer/hek293t_DMSO_polyAC_6_mer --kmer 6mer-CDS-AC.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_DMSO-1_polyAG_6_mer.txt,counts_10/hek293t_DMSO-2_polyAG_6_mer.txt -o counts_10_mean_kmer/hek293t_DMSO_polyAG_6_mer --kmer 6mer-CDS-AG.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_DMSO-1_polyCT_6_mer.txt,counts_10/hek293t_DMSO-2_polyCT_6_mer.txt -o counts_10_mean_kmer/hek293t_DMSO_polyCT_6_mer --kmer 6mer-CT.txt
cp counts_10/hek293t_*.txt counts_10_mean_kmer
## UTR
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_UTR_DMSO-1_polyAC_4_mer.txt,counts_10/hek293t_UTR_DMSO-2_polyAC_4_mer.txt -o counts_10_mean_kmer/hek293t_UTR_DMSO_polyAC_4_mer --kmer 4mer-5UTR-AC.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_UTR_DMSO-1_polyAG_4_mer.txt,counts_10/hek293t_UTR_DMSO-2_polyAG_4_mer.txt -o counts_10_mean_kmer/hek293t_UTR_DMSO_polyAG_4_mer --kmer 4mer-5UTR-AG.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_UTR_DMSO-1_polyCT_4_mer.txt,counts_10/hek293t_UTR_DMSO-2_polyCT_4_mer.txt -o counts_10_mean_kmer/hek293t_UTR_DMSO_polyCT_4_mer --kmer 4mer-CT.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_UTR_DMSO-1_polyAC_5_mer.txt,counts_10/hek293t_UTR_DMSO-2_polyAC_5_mer.txt -o counts_10_mean_kmer/hek293t_UTR_DMSO_polyAC_5_mer --kmer 5mer-5UTR-AC.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_UTR_DMSO-1_polyAG_5_mer.txt,counts_10/hek293t_UTR_DMSO-2_polyAG_5_mer.txt -o counts_10_mean_kmer/hek293t_UTR_DMSO_polyAG_5_mer --kmer 5mer-5UTR-AG.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_UTR_DMSO-1_polyCT_5_mer.txt,counts_10/hek293t_UTR_DMSO-2_polyCT_5_mer.txt -o counts_10_mean_kmer/hek293t_UTR_DMSO_polyCT_5_mer --kmer 5mer-CT.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_UTR_DMSO-1_polyAC_6_mer.txt,counts_10/hek293t_UTR_DMSO-2_polyAC_6_mer.txt -o counts_10_mean_kmer/hek293t_UTR_DMSO_polyAC_6_mer --kmer 6mer-5UTR-AC.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_UTR_DMSO-1_polyAG_6_mer.txt,counts_10/hek293t_UTR_DMSO-2_polyAG_6_mer.txt -o counts_10_mean_kmer/hek293t_UTR_DMSO_polyAG_6_mer --kmer 6mer-5UTR-AG.txt
python MeanRiboDensityAroundPolyPurineMotifsFiltered.py -i counts_10/hek293t_UTR_DMSO-1_polyCT_6_mer.txt,counts_10/hek293t_UTR_DMSO-2_polyCT_6_mer.txt -o counts_10_mean_kmer/hek293t_UTR_DMSO_polyCT_6_mer --kmer 6mer-CT.txt
- Calculate ratio between RocA and DMSO
## 02. calculate ratio
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyAC_4_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-003
_polyAC_4_mer.txt -o counts_10_mean_kmer/hek293t_RocA-003_polyAC_4_mer.ratio --kmer 4mer-CDS-AC.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyAC_4_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-03_polyAC_4_mer.txt -o counts_10_mean_kmer/hek293t_RocA-03_polyAC_4_mer.ratio --kmer 4mer-CDS-AC.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyAC_4_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-3_polyAC_4_mer.txt -o counts_10_mean_kmer/hek293t_RocA-3_polyAC_4_mer.ratio --kmer 4mer-CDS-AC.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyAG_4_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-003_polyAG_4_mer.txt -o counts_10_mean_kmer/hek293t_RocA-003_polyAG_4_mer.ratio --kmer 4mer-CDS-AG.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyAG_4_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-03_polyAG_4_mer.txt -o counts_10_mean_kmer/hek293t_RocA-03_polyAG_4_mer.ratio --kmer 4mer-CDS-AG.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyAG_4_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-3_polyAG_4_mer.txt -o counts_10_mean_kmer/hek293t_RocA-3_polyAG_4_mer.ratio --kmer 4mer-CDS-AG.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyCT_4_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-003_polyCT_4_mer.txt -o counts_10_mean_kmer/hek293t_RocA-003_polyCT_4_mer.ratio --kmer 4mer-CT.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyCT_4_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-03_polyCT_4_mer.txt -o counts_10_mean_kmer/hek293t_RocA-03_polyCT_4_mer.ratio --kmer 4mer-CT.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyCT_4_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-3_polyCT_4_mer.txt -o counts_10_mean_kmer/hek293t_RocA-3_polyCT_4_mer.ratio --kmer 4mer-CT.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyAC_5_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-003_polyAC_5_mer.txt -o counts_10_mean_kmer/hek293t_RocA-003_polyAC_5_mer.ratio --kmer 5mer-CDS-AC.txt
#python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyAC_5_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-03_polyAC_5_mer.txt -o counts_10_mean_kmer/hek293t_RocA-03_polyAC_5_mer.ratio --kmer 5mer-CDS-AC.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyAC_5_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-3_polyAC_5_mer.txt -o counts_10_mean_kmer/hek293t_RocA-3_polyAC_5_mer.ratio --kmer 5mer-CDS-AC.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyAG_5_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-003_polyAG_5_mer.txt -o counts_10_mean_kmer/hek293t_RocA-003_polyAG_5_mer.ratio --kmer 5mer-CDS-AG.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyAG_5_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-03_polyAG_5_mer.txt -o counts_10_mean_kmer/hek293t_RocA-03_polyAG_5_mer.ratio --kmer 5mer-CDS-AG.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyAG_5_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-3_polyAG_5_mer.txt -o counts_10_mean_kmer/hek293t_RocA-3_polyAG_5_mer.ratio --kmer 5mer-CDS-AG.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyCT_5_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-003_polyCT_5_mer.txt -o counts_10_mean_kmer/hek293t_RocA-003_polyCT_5_mer.ratio --kmer 5mer-CT.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyCT_5_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-03_polyCT_5_mer.txt -o counts_10_mean_kmer/hek293t_RocA-03_polyCT_5_mer.ratio --kmer 5mer-CT.txt
python CalRiboDensityRatioFilter.py -c counts_10_mean_kmer/hek293t_DMSO_polyCT_5_mer_mean.txt -t counts_10_mean_kmer/hek293t_RocA-3_polyCT_5_mer.txt -o counts_10_mean_kmer/hek293t_RocA-3_polyCT_5_mer.ratio --kmer 5mer-CT.txt
- plot the ratio values
# 03. plot ratio
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-003_polyAG_4_mer.ratio,counts_10_mean_kmer/hek293t_RocA-003_polyAC_4_mer.ratio -o counts_10_mean_kmer/RocA003_4_mer -t RocA003-AG,RocA003-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-03_polyAG_4_mer.ratio,counts_10_mean_kmer/hek293t_RocA-03_polyAC_4_mer.ratio -o counts_10_mean_kmer/RocA03_4_mer -t RocA03-AG,RocA03-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-3_polyAG_4_mer.ratio,counts_10_mean_kmer/hek293t_RocA-3_polyAC_4_mer.ratio -o counts_10_mean_kmer/RocA3_4_mer -t RocA3-AG,RocA3-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-003_polyAG_4_mer.ratio,counts_10_mean_kmer/hek293t_RocA-003_polyCT_4_mer.ratio -o counts_10_mean_kmer/RocA003_4_mer_AG_CT -t RocA003-AG,RocA003-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-03_polyAG_4_mer.ratio,counts_10_mean_kmer/hek293t_RocA-03_polyCT_4_mer.ratio -o counts_10_mean_kmer/RocA03_4_mer_AG_CT -t RocA03-AG,RocA03-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-3_polyAG_4_mer.ratio,counts_10_mean_kmer/hek293t_RocA-3_polyCT_4_mer.ratio -o counts_10_mean_kmer/RocA3_4_mer_AG_CT -t RocA3-AG,RocA3-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-003_polyAG_5_mer.ratio,counts_10_mean_kmer/hek293t_RocA-003_polyAC_5_mer.ratio -o counts_10_mean_kmer/RocA003_5_mer -t RocA003-AG,RocA003-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-03_polyAG_5_mer.ratio,counts_10_mean_kmer/hek293t_RocA-03_polyAC_5_mer.ratio -o counts_10_mean_kmer/RocA03_5_mer -t RocA03-AG,RocA03-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-3_polyAG_5_mer.ratio,counts_10_mean_kmer/hek293t_RocA-3_polyAC_5_mer.ratio -o counts_10_mean_kmer/RocA3_5_mer -t RocA3-AG,RocA3-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-003_polyAG_5_mer.ratio,counts_10_mean_kmer/hek293t_RocA-003_polyCT_5_mer.ratio -o counts_10_mean_kmer/RocA003_5_mer_AG_CT -t RocA003-AG,RocA003-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-03_polyAG_5_mer.ratio,counts_10_mean_kmer/hek293t_RocA-03_polyCT_5_mer.ratio -o counts_10_mean_kmer/RocA03_5_mer_AG_CT -t RocA03-AG,RocA03-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-3_polyAG_5_mer.ratio,counts_10_mean_kmer/hek293t_RocA-3_polyCT_5_mer.ratio -o counts_10_mean_kmer/RocA3_5_mer_AG_CT -t RocA3-AG,RocA3-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-003_polyAG_6_mer.ratio,counts_10_mean_kmer/hek293t_RocA-003_polyAC_6_mer.ratio -o counts_10_mean_kmer/RocA003_6_mer -t RocA003-AG,RocA003-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-03_polyAG_6_mer.ratio,counts_10_mean_kmer/hek293t_RocA-03_polyAC_6_mer.ratio -o counts_10_mean_kmer/RocA03_6_mer -t RocA03-AG,RocA03-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-3_polyAG_6_mer.ratio,counts_10_mean_kmer/hek293t_RocA-3_polyAC_6_mer.ratio -o counts_10_mean_kmer/RocA3_6_mer -t RocA3-AG,RocA3-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-003_polyAG_6_mer.ratio,counts_10_mean_kmer/hek293t_RocA-003_polyCT_6_mer.ratio -o counts_10_mean_kmer/RocA003_6_mer_AG_CT -t RocA003-AG,RocA003-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-03_polyAG_6_mer.ratio,counts_10_mean_kmer/hek293t_RocA-03_polyCT_6_mer.ratio -o counts_10_mean_kmer/RocA03_6_mer_AG_CT -t RocA03-AG,RocA03-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_RocA-3_polyAG_6_mer.ratio,counts_10_mean_kmer/hek293t_RocA-3_polyCT_6_mer.ratio -o counts_10_mean_kmer/RocA3_6_mer_AG_CT -t RocA3-AG,RocA3-CT -u 50 -d 50 --slide-window
## UTR
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-003_polyAG_4_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-003_polyAC_4_mer.ratio -o counts_10_mean_kmer/RocA003_UTR_4_mer -t RocA003-AG,RocA003-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-03_polyAG_4_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-03_polyAC_4_mer.ratio -o counts_10_mean_kmer/RocA03_UTR_4_mer -t RocA03-AG,RocA03-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-3_polyAG_4_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-3_polyAC_4_mer.ratio -o counts_10_mean_kmer/RocA3_UTR_4_mer -t RocA3-AG,RocA3-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-003_polyAG_4_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-003_polyCT_4_mer.ratio -o counts_10_mean_kmer/RocA003_UTR_4_mer_AG_CT -t RocA003-AG,RocA003-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-03_polyAG_4_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-03_polyCT_4_mer.ratio -o counts_10_mean_kmer/RocA03_UTR_4_mer_AG_CT -t RocA03-AG,RocA03-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-3_polyAG_4_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-3_polyCT_4_mer.ratio -o counts_10_mean_kmer/RocA3_UTR_4_mer_AG_CT -t RocA3-AG,RocA3-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-003_polyAG_5_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-003_polyAC_5_mer.ratio -o counts_10_mean_kmer/RocA003_UTR_5_mer -t RocA003-AG,RocA003-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-03_polyAG_5_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-03_polyAC_5_mer.ratio -o counts_10_mean_kmer/RocA03_UTR_5_mer -t RocA03-AG,RocA03-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-3_polyAG_5_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-3_polyAC_5_mer.ratio -o counts_10_mean_kmer/RocA3_UTR_5_mer -t RocA3-AG,RocA3-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-003_polyAG_5_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-003_polyCT_5_mer.ratio -o counts_10_mean_kmer/RocA003_UTR_5_mer_AG_CT -t RocA003-AG,RocA003-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-03_polyAG_5_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-03_polyCT_5_mer.ratio -o counts_10_mean_kmer/RocA03_UTR_5_mer_AG_CT -t RocA03-AG,RocA03-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-3_polyAG_5_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-3_polyCT_5_mer.ratio -o counts_10_mean_kmer/RocA3_UTR_5_mer_AG_CT -t RocA3-AG,RocA3-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-003_polyAG_6_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-003_polyAC_6_mer.ratio -o counts_10_mean_kmer/RocA003_UTR_6_mer -t RocA003-AG,RocA003-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-03_polyAG_6_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-03_polyAC_6_mer.ratio -o counts_10_mean_kmer/RocA03_UTR_6_mer -t RocA03-AG,RocA03-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-3_polyAG_6_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-3_polyAC_6_mer.ratio -o counts_10_mean_kmer/RocA3_UTR_6_mer -t RocA3-AG,RocA3-AC -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-003_polyAG_6_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-003_polyCT_6_mer.ratio -o counts_10_mean_kmer/RocA003_UTR_6_mer_AG_CT -t RocA003-AG,RocA003-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-03_polyAG_6_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-03_polyCT_6_mer.ratio -o counts_10_mean_kmer/RocA03_UTR_6_mer_AG_CT -t RocA03-AG,RocA03-CT -u 50 -d 50 --slide-window
python PlotRatio.py -i counts_10_mean_kmer/hek293t_UTR_RocA-3_polyAG_6_mer.ratio,counts_10_mean_kmer/hek293t_UTR_RocA-3_polyCT_6_mer.ratio -o counts_10_mean_kmer/RocA3_UTR_6_mer_AG_CT -t RocA3-AG,RocA3-CT -u 50 -d 50 --slide-window
- Calculate disome density around polypurine moitfs
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=~/Reference/human/hg38/ensemble
transcript=~/Reference/human/hg38/ensemble/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/test
attribute=$workdir/Disome_configure.txt
trans_info=$Ref/longest.transcripts.info.txt
up_trans=$workdir/374_up_genes_info.txt
down_trans=$workdir/643_down_genes_info.txt
# poly-purine
mkdir -p polymotifs/counts_10
## UTR
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_5UTR -M RPKM -u 100 -d 100 -l 0
-n 0 -F $transcript --kmer 4 --type 5UTR -S $down_trans --id-type transcript_id --base AG
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_5UTR -M RPKM -u 100 -d 100 -l 0
-n 0 -F $transcript --kmer 4 --type 5UTR -S $down_trans --id-type transcript_id --base AC
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_5UTR -M RPKM -u 100 -d 100 -l 0
-n 0 -F $transcript --kmer 4 --type 5UTR -S $down_trans --id-type transcript_id --base CT
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_5UTR -M RPKM -u 100 -d 100 -l 0
-n 0 -F $transcript --kmer 5 --type 5UTR -S $down_trans --id-type transcript_id --base AG
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_5UTR -M RPKM -u 100 -d 100 -l 0
-n 0 -F $transcript --kmer 5 --type 5UTR -S $down_trans --id-type transcript_id --base AC
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_5UTR -M RPKM -u 100 -d 100 -l 0
-n 0 -F $transcript --kmer 5 --type 5UTR -S $down_trans --id-type transcript_id --base CT
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_5UTR -M RPKM -u 100 -d 100 -l 0 -n 0 -F $transcript --kmer 6 --type 5UTR -S $down_trans --id-type transcript_id --base AG
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_5UTR -M RPKM -u 100 -d 100 -l 0 -n 0 -F $transcript --kmer 6 --type 5UTR -S $down_trans --id-type transcript_id --base AC
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_5UTR -M RPKM -u 100 -d 100 -l 0 -n 0 -F $transcript --kmer 6 --type 5UTR -S $down_trans --id-type transcript_id --base CT
## CDS
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_CDS -M RPKM -u 100 -d 100 -l 0 -n 0 -F $transcript --kmer 4 --type CDS -S $up_trans --id-type transcript_id --base AG
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_CDS -M RPKM -u 100 -d 100 -l 0 -n 0 -F $transcript --kmer 4 --type CDS -S $up_trans --id-type transcript_id --base AC
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_CDS -M RPKM -u 100 -d 100 -l 0 -n 0 -F $transcript --kmer 4 --type CDS -S $up_trans --id-type transcript_id --base CT
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_CDS -M RPKM -u 100 -d 100 -l 0 -n 0 -F $transcript --kmer 5 --type CDS -S $up_trans --id-type transcript_id --base AG
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_CDS -M RPKM -u 100 -d 100 -l 0 -n 0 -F $transcript --kmer 5 --type CDS -S $up_trans --id-type transcript_id --base AC
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_CDS -M RPKM -u 100 -d 100 -l 0 -n 0 -F $transcript --kmer 5 --type CDS -S $up_trans --id-type transcript_id --base CT
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_CDS -M RPKM -u 100 -d 100 -l 0 -n 0 -F $transcript --kmer 6 --type CDS -S $up_trans --id-type transcript_id --base AG
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_CDS -M RPKM -u 100 -d 100 -l 0 -n 0 -F $transcript --kmer 6 --type CDS -S $up_trans --id-type transcript_id --base AC
python RiboDensityAroundPolyPurineMotifs.py -f $attribute -c $trans_info -o polymotifs/counts_10/test_CDS -M RPKM -u 100 -d 100 -l 0 -n 0 -F $transcript --kmer 6 --type CDS -S $up_trans --id-type transcript_id --base CT
- Calculate ratio
python CalRiboDensityRatio.py counts_10/test_CDS_293FT-DMSO_polyAG_4_mer.txt counts_10/test_CDS_293FT-RocA03_polyAG_4_mer.txt counts_10/test_CDS_293FT_RocA03_polyAG_4_mer.ratio
python CalRiboDensityRatio.py counts_10/test_CDS_293FT-DMSO_polyAG_5_mer.txt counts_10/test_CDS_293FT-RocA03_polyAG_5_mer.txt counts_10/test_CDS_293FT_RocA03_polyAG_5_mer.ratio
python CalRiboDensityRatio.py counts_10/test_CDS_293FT-DMSO_polyAG_6_mer.txt counts_10/test_CDS_293FT-RocA03_polyAG_6_mer.txt counts_10/test_CDS_293FT_RocA03_polyAG_6_mer.ratio
python CalRiboDensityRatio.py counts_10/test_CDS_293FT-DMSO_polyAC_4_mer.txt counts_10/test_CDS_293FT-RocA03_polyAC_4_mer.txt counts_10/test_CDS_293FT_RocA03_polyAC_4_mer.ratio
python CalRiboDensityRatio.py counts_10/test_CDS_293FT-DMSO_polyAC_5_mer.txt counts_10/test_CDS_293FT-RocA03_polyAC_5_mer.txt counts_10/test_CDS_293FT_RocA03_polyAC_5_mer.ratio
python CalRiboDensityRatio.py counts_10/test_CDS_293FT-DMSO_polyAC_6_mer.txt counts_10/test_CDS_293FT-RocA03_polyAC_6_mer.txt counts_10/test_CDS_293FT_RocA03_polyAC_6_mer.ratio
python CalRiboDensityRatio.py counts_10/test_CDS_293FT-DMSO_polyCT_4_mer.txt counts_10/test_CDS_293FT-RocA03_polyCT_4_mer.txt counts_10/test_CDS_293FT_RocA03_polyCT_4_mer.ratio
python CalRiboDensityRatio.py counts_10/test_CDS_293FT-DMSO_polyCT_5_mer.txt counts_10/test_CDS_293FT-RocA03_polyCT_5_mer.txt counts_10/test_CDS_293FT_RocA03_polyCT_5_mer.ratio
python CalRiboDensityRatio.py counts_10/test_CDS_293FT-DMSO_polyCT_6_mer.txt counts_10/test_CDS_293FT-RocA03_polyCT_6_mer.txt counts_10/test_CDS_293FT_RocA03_polyCT_6_mer.ratio
- Plot ratio
# 03. plot ratio
python PlotRatio.py -i counts_10/test_CDS_293FT_RocA03_polyAG_4_mer.ratio,counts_10/test_CDS_293FT_RocA03_polyAC_4_mer.ratio -o counts_10/test_CDS_293FT_4mer_AG_AC -t RocA03-AG,RocA03-AC -u 100 -d 100 --slide-window
python PlotRatio.py -i counts_10/test_CDS_293FT_RocA03_polyAG_4_mer.ratio,counts_10/test_CDS_293FT_RocA03_polyCT_4_mer.ratio -o counts_10/test_CDS_293FT_4mer_AG_CT -t RocA03-AG,RocA03-CT -u 100 -d 100 --slide-window
python PlotRatio.py -i counts_10/test_CDS_293FT_RocA03_polyAG_5_mer.ratio,counts_10/test_CDS_293FT_RocA03_polyAC_5_mer.ratio -o counts_10/test_CDS_293FT_5mer_AG_AC -t RocA03-AG,RocA03-AC -u 100 -d 100 --slide-window
python PlotRatio.py -i counts_10/test_CDS_293FT_RocA03_polyAG_5_mer.ratio,counts_10/test_CDS_293FT_RocA03_polyCT_5_mer.ratio -o counts_10/test_CDS_293FT_5mer_AG_CT -t RocA03-AG,RocA03-CT -u 100 -d 100 --slide-window
python PlotRatio.py -i counts_10/test_CDS_293FT_RocA03_polyAG_6_mer.ratio,counts_10/test_CDS_293FT_RocA03_polyAC_6_mer.ratio -o counts_10/test_CDS_293FT_6mer_AG_AC -t RocA03-AG,RocA03-AC -u 100 -d 100 --slide-window
python PlotRatio.py -i counts_10/test_CDS_293FT_RocA03_polyAG_6_mer.ratio,counts_10/test_CDS_293FT_RocA03_polyCT_6_mer.ratio -o counts_10/test_CDS_293FT_6mer_AG_CT -t RocA03-AG,RocA03-CT -u 100 -d 100 --slide-window
- Metagene analysis via RiboMiner
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA
attribute=$workdir/GSE102720_configure.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='Naive-DMSO,Naive-RocA03,Aglaiacized-DMSO,Aglaiacized-RocA03'
replicates='Naive-DMSO-1,Naive-DMSO-2__Naive-RocA03__Aglaiacized-DMSO-1,Aglaiacized-DMSO-2__Aglaiacized-RocA03'
mkdir -p $results
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_RocA03_up_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 0 -e 5 --norm no -y 100 --CI 0.95 --type UTR -S 374_up_genes_info.txt
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_RocA03_down_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 0 -e 5 --norm no -y 100 --CI 0.95 --type UTR -S 643_down_genes_info.txt
PlotMetageneAnalysis -i $results/MA_RocA03_up_unnormed_dataframe.txt -o $results/MA_RocA03_up_unnormed -g $groups -r $replicates -u 100 -d 400 --mode mean
PlotMetageneAnalysis -i $results/MA_RocA03_down_unnormed_dataframe.txt -o $results/MA_RocA03_down_unnormed -g $groups -r $replicates -u 100 -d 400 --mode mean
This is similar to Figure 4C and Figure 4D
- configure files for 4 tumor cell samples
$ cat ./data/attributes_375.txt
bamFiles readLengths Offsets bamLegends
../07.STAR/SRR11542147_STAR/SRR11542147.Aligned.toTranscriptome.out.sorted.bam 26,27,28,29,30,31 12,12,12,12,12,13 DMSO-1-375
../07.STAR/SRR11542148_STAR/SRR11542148.Aligned.toTranscriptome.out.sorted.bam 26,27,28,29,30,31 12,12,12,12,12,13 DMSO-2-375
../07.STAR/SRR11542155_STAR/SRR11542155.Aligned.toTranscriptome.out.sorted.bam 25,26,27,28,29,30,31 12,12,12,12,12,12,13 RocA03-375
../07.STAR/SRR11542156_STAR/SRR11542156.Aligned.toTranscriptome.out.sorted.bam 25,26,27,28,29,30,31 12,12,12,12,12,12,13 RocA3-375
$ cat ./data/attributes_520.txt
bamFiles readLengths Offsets bamLegends
../07.STAR/SRR11542143_STAR/SRR11542143.Aligned.toTranscriptome.out.sorted.bam 25,26,27,28,29,30,31 12,12,12,12,12,12,13 DMSO-1-520
../07.STAR/SRR11542144_STAR/SRR11542144.Aligned.toTranscriptome.out.sorted.bam 25,26,27,28,29,30,31 12,12,12,12,12,12,13 DMSO-2-520
../07.STAR/SRR11542151_STAR/SRR11542151.Aligned.toTranscriptome.out.sorted.bam 25,26,27,28,29,30,31 12,12,12,12,12,12,13 RocA03-520
../07.STAR/SRR11542152_STAR/SRR11542152.Aligned.toTranscriptome.out.sorted.bam 25,26,27,28,29,30,31 12,12,12,12,12,12,13 RocA3-520
$ cat ./data/attributes_936.txt
bamFiles readLengths Offsets bamLegends
../07.STAR/SRR11542145_STAR/SRR11542145.Aligned.toTranscriptome.out.sorted.bam 26,27,28,29,30,31 12,12,12,12,12,13 DMSO-1-936
../07.STAR/SRR11542146_STAR/SRR11542146.Aligned.toTranscriptome.out.sorted.bam 26,27,28,29,30,31 12,12,12,12,12,13 DMSO-2-936
../07.STAR/SRR11542153_STAR/SRR11542153.Aligned.toTranscriptome.out.sorted.bam 26,27,28,29,30,31 12,12,12,12,12,13 RocA03-936
../07.STAR/SRR11542154_STAR/SRR11542154.Aligned.toTranscriptome.out.sorted.bam 27,28,29,30,31 12,12,12,12,13 RocA3-936
$ cat ./data/attributes_1650.txt
bamFiles readLengths Offsets bamLegends
../07.STAR/SRR11542141_STAR/SRR11542141.Aligned.toTranscriptome.out.sorted.bam 30,31,32,33,34 12,13,13,13,13 DMSO-1-1650
../07.STAR/SRR11542142_STAR/SRR11542142.Aligned.toTranscriptome.out.sorted.bam 30,31,32,33,34 12,13,13,13,13 DMSO-2-1650
../07.STAR/SRR11542149_STAR/SRR11542149.Aligned.toTranscriptome.out.sorted.bam 30,31,32,33,34 12,13,13,13,13 RocA03-1650
../07.STAR/SRR11542150_STAR/SRR11542150.Aligned.toTranscriptome.out.sorted.bam 30,31,32,33,34 12,13,13,13,13 RocA3-1650
- Metagene analysis via RiboMiner
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA_375
attribute=$workdir/attributes_375.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='DMSO-375,RocA03-375,RocA3-375'
replicates='DMSO-1-375,DMSO-2-375__RocA03-375__RocA3-375'
mkdir -p $results
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_375_RocA03-down_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 0 -e 5 --norm no -y 100 --CI 0.95 --type UTR -S 643_down_genes_info.txt
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_375_RocA03-up_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 0 -e 5 --norm no -y 100 --CI 0.95 --type UTR -S 374_up_genes_info.txt
PlotMetageneAnalysis -i $results/MA_375_RocA03-down_unnormed_dataframe.txt -o $results/MA_375_unnormed_RocA03-down -g $groups -r $replicates -u 100 -d 400 --mode mean
PlotMetageneAnalysis -i $results/MA_375_RocA03-up_unnormed_dataframe.txt -o $results/MA_375_unnormed_RocA03-up -g $groups -r $replicates -u 100 -d 400 --mode mean
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA_520
attribute=$workdir/attributes_520.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='DMSO-520,RocA03-520,RocA3-520'
replicates='DMSO-1-520,DMSO-2-520__RocA03-520__RocA3-520'
mkdir -p $results
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_520_RocA03-down_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 0 -e 5 --norm no -y 100 --CI 0.95 --type UTR -S 643_down_genes_info.txt
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_520_RocA03-up_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 0 -e 5 --norm no -y 100 --CI 0.95 --type UTR -S 374_up_genes_info.txt
PlotMetageneAnalysis -i $results/MA_520_RocA03-down_unnormed_dataframe.txt -o $results/MA_520_unnormed_RocA03-down -g $groups -r $replicates -u 100 -d 400 --mode mean
PlotMetageneAnalysis -i $results/MA_520_RocA03-up_unnormed_dataframe.txt -o $results/MA_520_unnormed_RocA03-up -g $groups -r $replicates -u 100 -d 400 --mode mean
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA_936
attribute=$workdir/attributes_936.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='DMSO-936,RocA03-936,RocA3-936'
replicates='DMSO-1-936,DMSO-2-936__RocA03-936__RocA3-936'
mkdir -p $results
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_936_RocA03-down_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 0 -e 5 --norm no -y 100 --CI 0.95 --type UTR -S 643_down_genes_info.txt
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_936_RocA03-up_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 0 -e 5 --norm no -y 100 --CI 0.95 --type UTR -S 374_up_genes_info.txt
PlotMetageneAnalysis -i $results/MA_936_RocA03-down_unnormed_dataframe.txt -o $results/MA_936_unnormed_RocA03-down -g $groups -r $replicates -u 100 -d 400 --mode mean
PlotMetageneAnalysis -i $results/MA_936_RocA03-up_unnormed_dataframe.txt -o $results/MA_936_unnormed_RocA03-up -g $groups -r $replicates -u 100 -d 400 --mode mean
workdir=`pwd`
BamDir=$workdir/../07.STAR
Ref=/workdata/home/lifj/lifj/data/Reference/human
transcript=/workdata/home/lifj/lifj/data/Reference/human/RiboCode_annot_Human/RiboCode_annot/transcripts_sequence.fa
results=$workdir/MA_1650
attribute=$workdir/attributes_1650.txt
trans_info=$Ref/longest.transcripts.info.txt
groups='DMSO-1650,RocA03-1650,RocA3-1650'
replicates='DMSO-1-1650,DMSO-2-1650__RocA03-1650__RocA3-1650'
mkdir -p $results
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_1650_RocA03-down_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 0 -e 5 --norm no -y 100 --CI 0.95 --type UTR -S 643_down_genes_info.txt
MetageneAnalysis -f $attribute -c $trans_info -o $results/MA_1650_RocA03-up_unnormed -U codon -M RPKM -u 100 -d 400 -l 0 -n 0 -m 0 -e 5 --norm no -y 100 --CI 0.95 --type UTR -S 374_up_genes_info.txt
PlotMetageneAnalysis -i $results/MA_1650_RocA03-down_unnormed_dataframe.txt -o $results/MA_1650_unnormed_RocA03-down -g $groups -r $replicates -u 100 -d 400 --mode mean
PlotMetageneAnalysis -i $results/MA_1650_RocA03-up_unnormed_dataframe.txt -o $results/MA_1650_unnormed_RocA03-up -g $groups -r $replicates -u 100 -d 400 --mode mean
The methods are the same as that for Figure 4C and Figure 4D
RDGs=IRGs RUGs=ERGs