Scripts to run AlphaFold3, Boltz-1 and Chai-1 with MMseqs2 Multiple sequence alignments (MSAs) and custom templates.
We recommend installing this package in a virtual environment or conda / micromamba environment. Python 3.11 is recommended, but the package should work with Python 3.9 and above.
To set up a conda/micromamba environment, run:
conda env create abcfold python=3.11
conda activate abcfoldor
micromamba env create abcfold python=3.11
micromamba activate abcfoldTo install the package from PyPI, run:
python -m pip install abcfoldOr, to install the package from source, first clone the repository and then run:
python -m pip install .If you wish to help develop this package, you can install the development dependencies by running:
python -m pip install -e .
python -m pip install -r requirements-dev.txt
python -m pre-commit installABCFold will run Alphafold3, Boltz-1 and Chai-1 consecutively. The program takes an input of a JSON in the Alphafold3 format (For full instruction on how to format this, click here). An example JSON is shown below:
{
"name": "2PV7",
"sequences": [
{
"protein": {
"id": ["A", "B"],
"sequence": "GMRESYANENQFGFKTINSDIHKIVIVGGYGKLGGLFARYLRASGYPISILDREDWAVAESILANADVVIVSVPINLTLETIERLKPYLTENMLLADLTSVKREPLAKMLEVHTGAVLGLHPMFGADIASMAKQVVVRCDGRFPERYEWLLEQIQIWGAKIYQTNATEHDHNMTYIQALRHFSTFANGLHLSKQPINLANLLALSSPIYRLELAMIGRLFAQDAELYADIIMDKSENLAVIETLKQTYDEALTFFENNDRQGFIDAFHKVRDWFGDYSEQFLKESRQLLQQANDLKQG"
}
}
],
"modelSeeds": [1],
"dialect": "alphafold3",
"version": 1
}Please make sure you have AlphaFold3 installed on your system (Instructions here) and have procured the model parameters. Boltz-1 and Chai-1 are installed upon runtime.
For the majority of jobs, ABCFold can be run as follows:
abcfold <input_json> <output_dir> -abc --mmseqs2 --model_params <path_to_af3_model_params>Note
--model_params is stored after the first run, therefore subsequent ABCFold jobs don't require this flag.
Note
If you wish to run ABCFold with the AlphaFold3 JACKHMMER MSA search, you need to remove the --mmseqs2 flag and provide the --database flag with the path to the directory containing the AlphaFold3 databases.
The --database path will also be stored after the first run and won't be required in subsequent ABCFold jobs.
Warning
--model_params and --database will need to be provided again if you do a fresh install.
However, there you may wish to use the following flags to add run time options such as the use of templates, or the number of models to create.
<input_json>: Path to the input AlphaFold3 JSON file.<output_dir>: Path to the output directory.-a,-b,-c(--alphafold3,--boltz1,--chai1): Flags to run Alphafold3, Boltz-1 and Chai-1 respectively. If none of these flags are provided, Alphafold3 will be run by default.--mmseqs2: [optional] Flag to use MMseqs2 MSAs and templates.--override: [optional] Flag to override the existing output directory.--save_input: [optional] Flag to save the input JSON file in the output directory.
--model_params: Path to the directory containing the AlphaFold3 model parameters.--database: [optional] Path to the directory containing the AlphaFold3 databases #Note: This is not used if using the--mmseqs2flag.--use_af3_template_search[optional] If providing your own custom MSA or you've ran--mmseqs, allow Alphafold3 to search for templates
-
--num_templates: [optional] The number of templates to use (default: 20) -
--custom_template: [optional] Path to a custom template file in mmCIF format or a list of custom templates. A more detailed decription on how to use the custom template argument can be found below Visualisation arguments. -
--custom_template_chain: [conditionally required] The chain ID of the chain to use in your custom template, only required if using a multi-chain template. If providing a list of custom templates, you will need to provide a list of custom template chains. -
--target_id: [conditionally required] The ID of the sequence the custom template relates to, only required if modelling a complex. If providing a list of custom templates, you can provide a single target ID if they all relate to the same target. Otherwise, you should provide a list of target IDs corresponding to the list of custom templates.
--no_server: [optional] Flag to not run the server for the output page (see below) but the pages are created , useful for running on a cluster.--no_visuals: [optional] Flag to not generate any output pages or PAE plots and only output the models.
If you wanted to provide a custom template, custom_a.pdb for your protein sequence with the ID A and you have your template has two chains: chain A and chain B and chain B is what you want the template to be, you could run:
abcfold <input_json> <output_dir> -abc --mmseqs2 --custom_template custom_a.pdb --custom_template_chain B --target_id A
If you had multiple IDs in your input sequence, multiple template files and you wanted to provide 3 custom templates, chain A from custom_a.pdb, chain B from custom_b.pdb, and chain B from custom_c.pdb, where custom_a.pdb and custom_b.pdb correspond to the ID A and custom_c.pdb corresponds to the ID B, you could run:
abcfold <input_json> <output_dir> -abc --mmseqs2 --custom_template custom_a.pdb custom_b.pdb custom_c.pdb --custom_template_chain A B B --target_id A A B
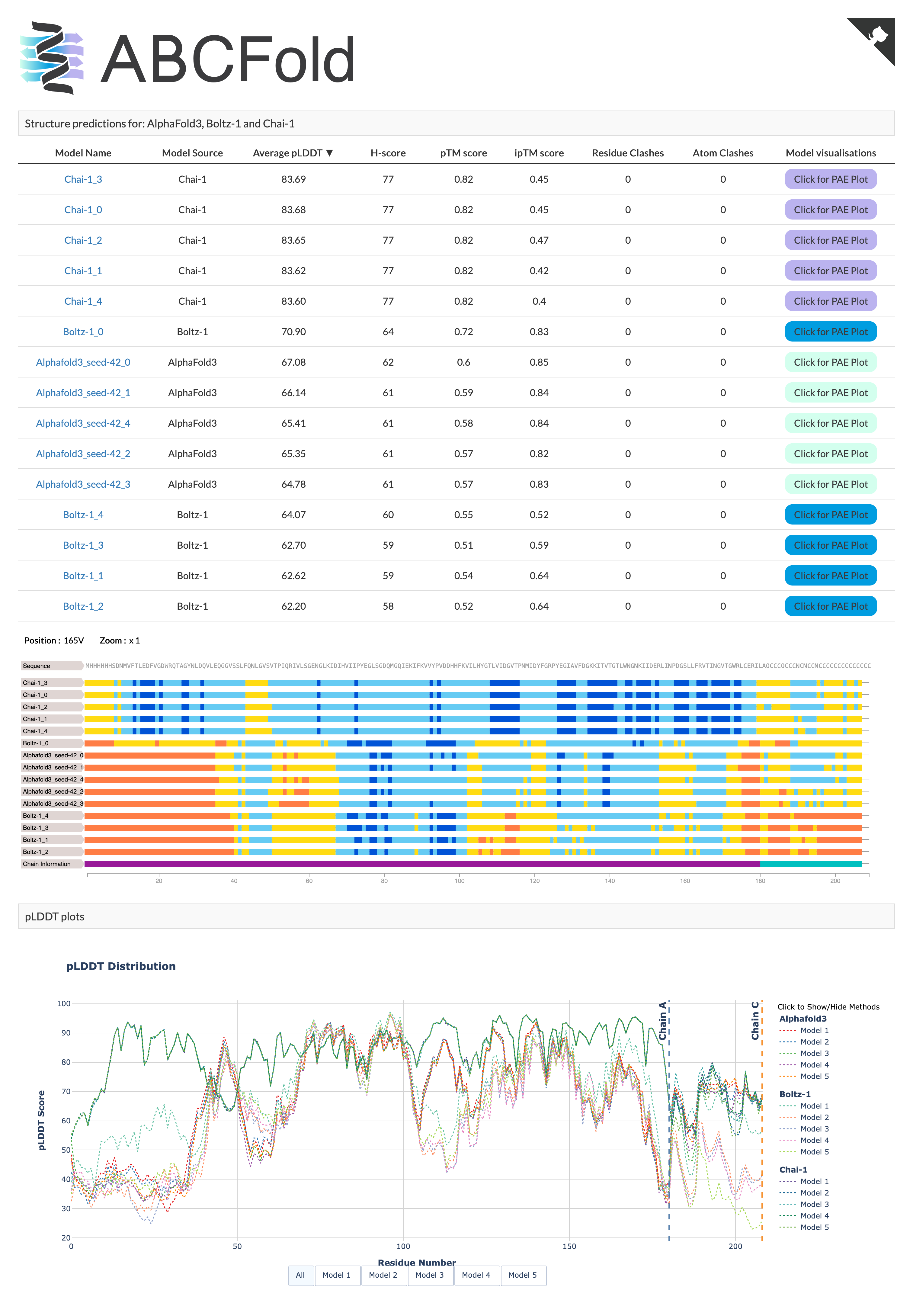
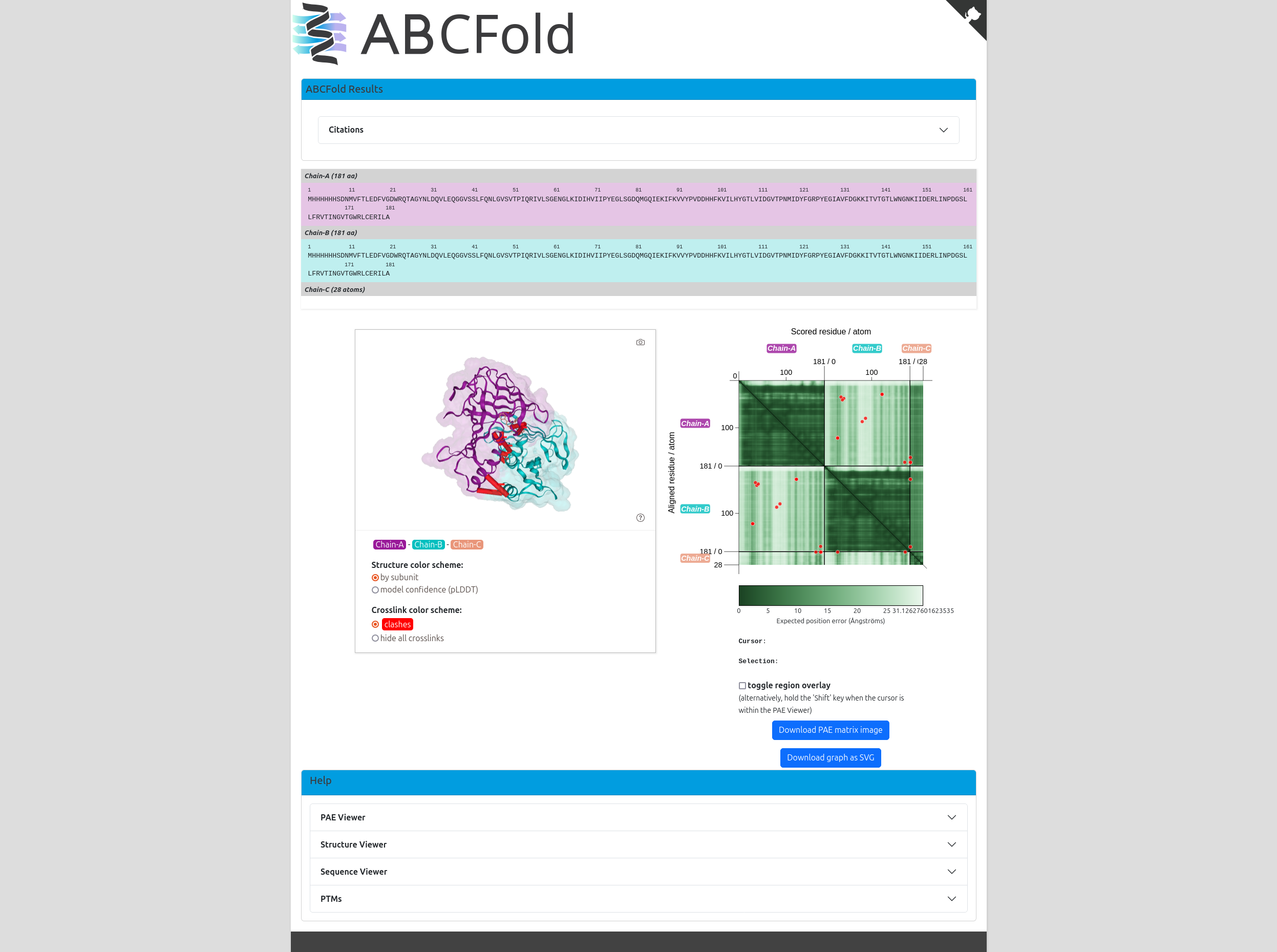
ABCFold will output the AlphaFold, Boltz and/or Chai models in the <output_dir>, it will also produce an output page containing a results table and informative PAE viewer. This is opened automatically in your default browser unless the --no_server or --no_visuals flags are used.
Unless the --no_visuals flag is used, you can then open the output pages by running:
python <output_dir>/open_output.pyThe output page will be available on http://localhost:8000/index.html. If you need to rerun the server to create the output,
you will find open_output.py in your <output_dir>. This needs to be run from your <output_dir>.
Below are scripts for adding MMseqs2 MSAs and custom templates to AlphaFold3 input JSON files.
Warning
These scripts will only modify the input JSON files, I.E. they will NOT run AlphaFold3, Boltz-1 and Chai-1.
To add MMseqs2 MSAs and templates to the AlphaFold3 input JSON, you can use the mmseqs2msa:
To run the script with templates, use the following command:
mmseqs2msa --input_json <input_json> --output_json <output_json> --templates --num_templates <num_templates><input_json>: Path to the input AlphaFold3 JSON file.<output_json>: [optional] Path to the output JSON file (default:<input_json_stem>_mmseqs.json).<num_templates>: [optional] The number of templates to use (default: 20)
To run the script without templates, use the following command:
mmseqs2msa --input_json <input_json> --output_json <output_json><input_json>: Path to the input AlphaFold3 JSON file.<output_json>: [optional] Path to the output JSON file (default:<input_json_stem>_mmseqs.json).
You may wish to add custom templates to your AlphaFold3 job, e.g. homologues which have yet to be deposited in the PDB. You can do so in two ways:
If you just wish to add a custom template, you can use custom_templates:
custom_templates --input_json <input_json> --output_json <output_json> --custom_template <custom_template> --custom_template_chain <custom_template_chain> --target_id <target_id><input_json>: Path to the input AlphaFold3 JSON file.<output_json>: [optional] Path to the output JSON file (default:<input_json_stem>_custom_template.json).<custom_template>: [optional] Path to a custom template file in mmCIF format or a list of custom templates.<custom_template_chain>: [conditionally required] The chain ID of the chain to use in your custom template, only required if using a multi-chain template. If providing a list of custom templates, you will need to provide a list of custom template chains.<target_id>: [conditionally required] The ID of the sequence the custom template relates to, only required if modelling a complex. If providing a list of custom templates, you can provide a single target ID if they all relate to the same target. Otherwise, you should provide a list of target IDs corresponding to the list of custom templates.
If you wish to add a custom template and generate an MMseqs2 MSA/templates, you can use mmseqs2msa:
mmseqs2msa --input_json <input_json> --output_json <output_json> --templates --num_templates <num_templates> --custom_template <custom_template> --custom_template_chain <custom_template_chain> --target_id <target_id><input_json>: Path to the input AlphaFold3 JSON file.<output_json>: [optional] Path to the output JSON file (default:<input_json_stem>_mmseqs.json).<num_templates>: [optional] The number of templates to use (default: 20)<custom_template>: [optional] Path to a custom template file in mmCIF format or a list of custom templates.<custom_template_chain>: [conditionally required] The chain ID of the chain to use in your custom template, only required if using a multi-chain template. If providing a list of custom templates, you will need to provide a list of custom template chains.<target_id>: [conditionally required] The ID of the sequence the custom template relates to, only required if modelling a complex. If providing a list of custom templates, you can provide a single target ID if they all relate to the same target. Otherwise, you should provide a list of target IDs corresponding to the list of custom templates.
Below is an example of a hetero-3-mer. When modelling a homo-oligomer, id is given as a list, you should select 1 of the identifiers in the list.
{
"name": "7ZYH",
"sequences": [
{
"protein": {
"id": "A",
"sequence": "SNAESKIKDCPWYDRGFCKHGPLCRHRHTRRVICVNYLVGFCPEGPSCKFMHPRFELPMGTTEQ"
}
},
{
"protein": {
"id": ["B", "C"],
"sequence": "SNAGSINGVPLLEVDLDSFEDKPWRKPGADLSDYFNYGFNEDTWKAYCEKQKRIRMGLEVIPVTSTTNK"
}
}
],
"modelSeeds": [1],
"dialect": "alphafold3",
"version": 1
}If you want to add a custom template to the first sequence, you can use --target_id A. If you wish to add a custom template to the second sequence, use --target_id B or --target_id C.
Contributions are welcome! Please open an issue or submit a pull request.