Mutational signature analysis is very active and important area of interest. There are several packages available now for mutational signature analysis and they all use different approaches and give nontrivially different results. Because of the differences in their results, it is important for researchers to survey the available tools and make choose the one that best suits their application. There is a need for software that can aggregate the results from different packages and present them in a user friendly way so as to facilitate effective comparison.
We created this package MetaMutationalSigs to facilitate comprehensive mutational signature analysis by creating a wrapper for different packages and providing a standard format for their outputs so that they can be effectively compared. We have also standardized the input formats accepted by various packages so ease interoperability. We also create standard visualizations for the results of all packages to ensure easy analysis. Our software is easy to install and use through Docker ,a package manager that automates the dependencies.
If you have questions, you can contact the author, Palash Pandey at pp535@drexel.edu OR PI Gail Rosen at eesi.pogo@gmail.com
docker pull pp535/metamutationalsigs
The docker image can be found at dockerhub here: https://hub.docker.com/r/pp535/metamutationalsigs
VCF files.
To run metamutationalsigs without using sigflow and sigfit on the data from your VCF file directory C:\Users\...full_path...\docker_input_test. Just replace C:\Users\...full_path...\docker_input_test/ with absolute path to your input directory that has VCF files. The results will be in a zipped file in your input directory.
docker run --rm -it -v C:\Users\...full_path...\docker_input_test/:/app/input_vcf_dir pp535/metamutationalsigs --i "input_vcf_dir" --sigflow --sigfit
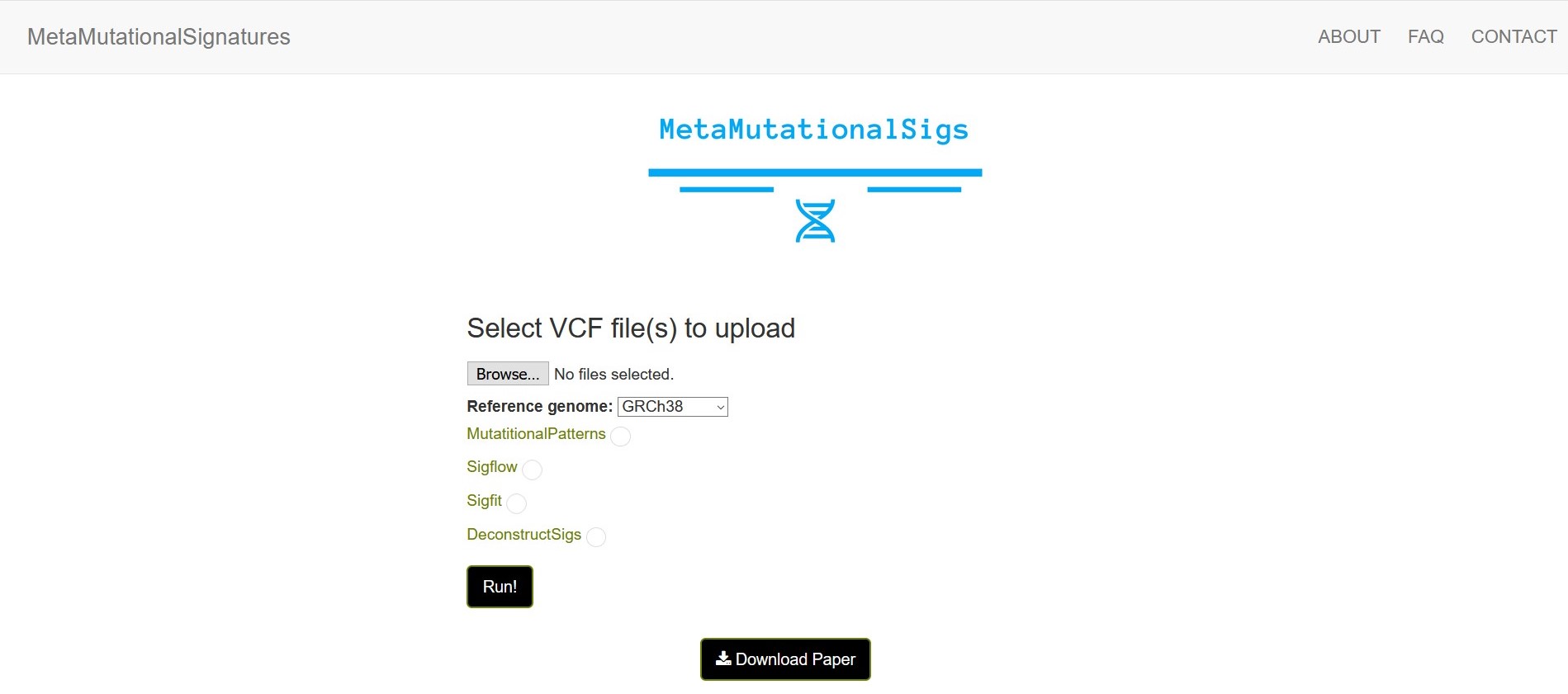
We have browser UI available as well:
docker run --rm -it -p 5001:5001 -v C:\Users\...full_path...\docker_input_test/:/app/input_vcf_dir pp535/metamutationalsigs --browser
Just replace C:\Users\...full_path...\docker_input_test/ with absolute path to your input directory that has VCF files. Then go to your browser at http://localhost:5001/ for the browser user interface.
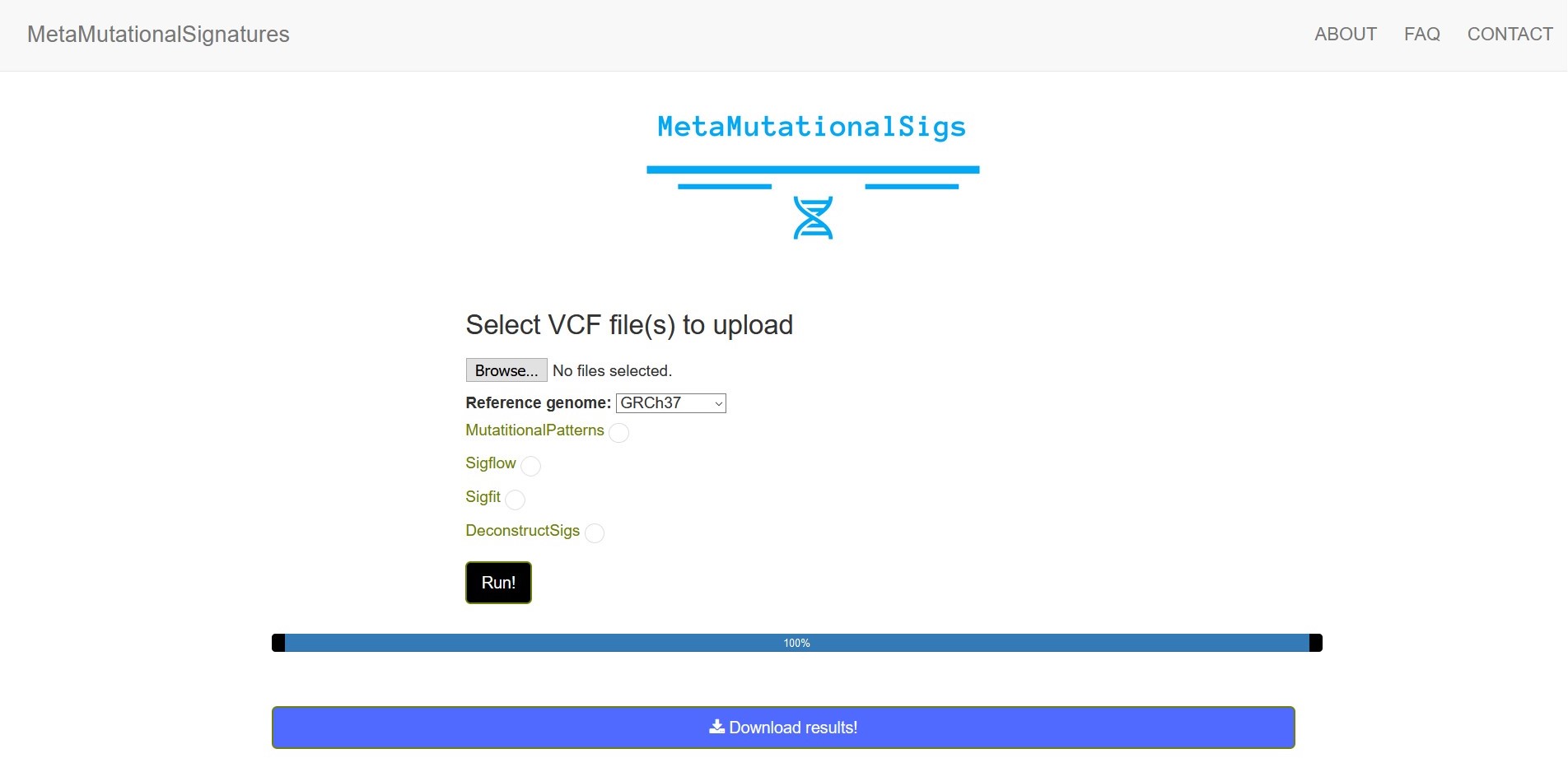
Once you select your VCF file directory and the tools that you would like to run, you will see a progress bar and when the progress bar reaches 100%, you can download the results as a zip file using the download results button.
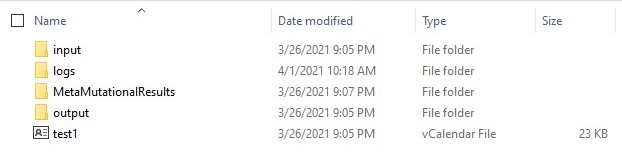
The output is returned as a compressed directory called MetaMutationalResults. Once uncompressed, this looks below. Directory MetaMutationalResults has the relevant results.
Inside MetaMutationalResults, we can find a folder for each tool that was selected.
Here is a summary of the files generated:
| File Name | Format | Description |
|---|---|---|
| Heatmap_contributions_all_sigs_legacy.pdf | Contributions for all COSMIC Legacy SBS signatures to the overall signature. | |
| Heatmap_contributions_all_sigs_SBS.pdf | Contributions for all COSMIC V3 SBS signatures. | |
| Heatmap_COSMIC_legacy.pdf | Heatmap for difference between the predicted contributions by different tools. One for each sample. | |
| Heatmap_COSMIC_V3.pdf | Heatmap for difference between the predicted contributions by different tools. One for each sample. | |
| legacy_pie_charts.html | html | Interactive pie charts of COSMIC legacy SBS contribution, per sample and for each tool. |
| sbs_pie_charts.html | html | Interactive pie charts of COSMIC V3 SBS signature contributions, per sample and for each tool. |
| legacy_rmse_bar_plot.pdf | Reconstruction error using COSMIC Legacy SBS signatures for each tool. | |
| sbs_rmse_bar_plot.png | Reconstruction error using COSMIC V3 SBS signatures for each tool. | |
| toolname_results\legacy_sample_error.csv | csv | Data used to create the bar plot. |
| toolname_results\legacy_sample_contribution.csv | csv | Data used to create heatmap and pie chart. |
| toolname_results\sbs_sample_error.csv | csv | Data used to create the bar plot. |
| toolname_results\sbs_sample_contribution.csv | csv | Data used to create heatmap and pie chart. |
- MutatitionalPatterns https://bioconductor.org/packages/release/bioc/html/MutationalPatterns.html
- Sigflow/ Sigminer https://github.com/ShixiangWang/sigflow
- Sigfit https://github.com/kgori/sigfit
- DeconstructSigs https://github.com/raerose01/deconstructSigs
Omichessan, H., Severi, G., & Perduca, V. (2019). Computational tools to detect signatures of mutational processes in DNA from tumours: A review and empirical comparison of performance. PLOS ONE, 14(9), e0221235. https://doi.org/10.1371/journal.pone.0221235
MetamutationalSigs supports:
GRCh37/ hg19 Homo sapiens
GRCh38/ hg38 Homo sapiens
GRCm37/ mm9 Mus musculus
GRCm38.p6/ mm10 Mus musculus
Rnor_6.0/ rn6 Rattus norvegicus
Your files need to be in VCF format. For more information https://www.internationalgenome.org/wiki/Analysis/vcf4.0/
All analysis is run locally. No data leaves your computer. The web browser user interface is also running locally on your computer, so you can feel free to analyze your protected data.
- V1 - COSMIC reference signatures V3.1 June 2020
- V2* Current - COSMIC reference signatures updated to V3.2 March 2020