An open source Python library for automated feature engineering based on Genetic Programming.
- Free software: BSD license
- Documentation: https://evolutionary-forest.readthedocs.io
Feature engineering is a long-standing issue that has plagued machine learning practitioners for many years. Deep learning techniques have significantly reduced the need for manual feature engineering in recent years. However, a critical issue is that the features discovered by deep learning methods are difficult to interpret.
In the domain of interpretable machine learning, genetic programming has demonstrated to be a promising method for automated feature construction, as it can improve the performance of traditional machine learning systems while maintaining similar interpretability. Nonetheless, such a potent method is rarely mentioned by practitioners. We believe that the main reason for this phenomenon is that there is still a lack of a mature package that can automatically build features based on the genetic programming algorithm. As a result, we propose this package with the goal of providing a powerful feature construction tool for enhancing existing state-of-the-art machine learning algorithms, particularly decision-tree-based algorithms.
- A powerful feature construction tool for generating interpretable machine learning features.
- A reliable machine learning model with powerful performance on small datasets.
pip install -U evolutionary_forestpip install git+https://github.com/hengzhe-zhang/EvolutionaryForest.git- Evolutionary Forest: Ensemble GP for Decision Trees (TEVC 2021)
- SR-Forest: Ensemble GP for Decision Trees + Linear Models (TEVC 2023)
- SHM-GP: Semantic Host Mutation (TEVC 2024)
- MMTGP: Modular Multi-Tree GP (TEVC 2024)
- RAG-SR: Retrieve-Augmented Deep Symbolic Regression (ICLR 2025)
An example of usage:
X, y = load_diabetes(return_X_y=True)
x_train, x_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
r = EvolutionaryForestRegressor(max_height=3, normalize=True, select='AutomaticLexicase',
gene_num=10, boost_size=100, n_gen=20, n_pop=200, cross_pb=1,
base_learner='Random-DT', verbose=True)
r.fit(x_train, y_train)
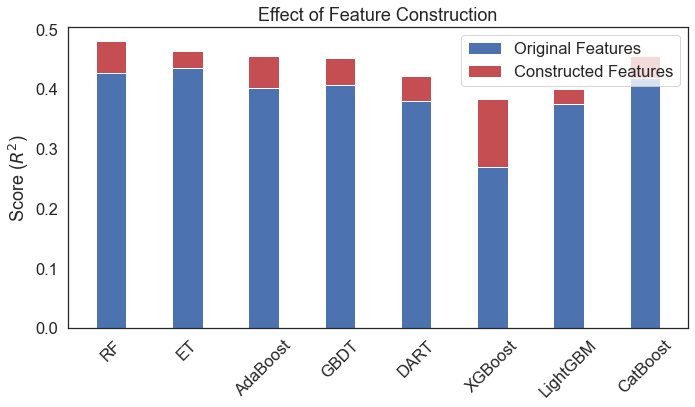
print(r2_score(y_test, r.predict(x_test)))An example of improvements brought about by constructed features:
Here are some notebook examples of using Evolutionary Forest:
Tutorial: English Version | 中文版本
This package was created with Cookiecutter and the audreyr/cookiecutter-pypackage project template.
Please cite our paper if you find it helpful:
@article{zhang2021evolutionary,
title = {An Evolutionary Forest for Regression},
author = {Zhang, Hengzhe and Zhou, Aimin and Zhang, Hu},
journal = {IEEE Transactions on Evolutionary Computation},
volume = {26},
number = {4},
pages = {735--749},
year = {2021},
publisher = {IEEE}
}
@article{zhang2023sr,
title = {SR-Forest: A Genetic Programming based Heterogeneous Ensemble Learning Method},
author = {Zhang, Hengzhe and Zhou, Aimin and Chen, Qi and Xue, Bing and Zhang, Mengjie},
journal = {IEEE Transactions on Evolutionary Computation},
year = {2023},
publisher = {IEEE}
}
@article{zhang2023semantic,
title = {A Semantic-Based Hoist Mutation Operator for Evolutionary Feature Construction in Regression},
author = {Zhang, Hengzhe and Chen, Qi and Xue, Bing and Banzhaf, Wolfgang and Zhang, Mengjie},
journal = {IEEE Transactions on Evolutionary Computation},
year = {2023},
publisher = {IEEE}
}
@article{zhang2023modular,
title = {Modular Multi-Tree Genetic Programming for Evolutionary Feature Construction for Regression},
author = {Zhang, Hengzhe and Chen, Qi and Xue, Bing and Banzhaf, Wolfgang and Zhang, Mengjie},
journal = {IEEE Transactions on Evolutionary Computation},
year = {2023},
publisher = {IEEE}
}