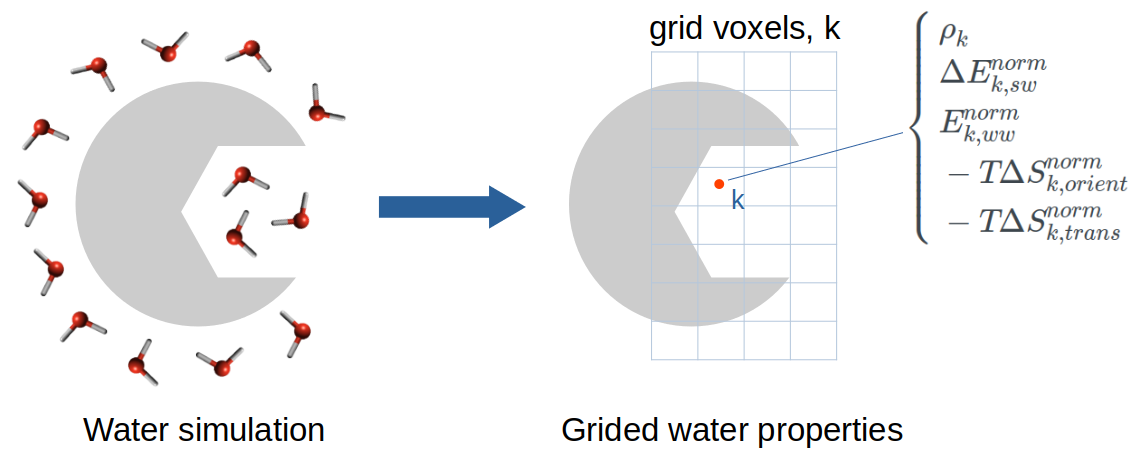
- BTK inhibitor: water replacement
- PDE10A inhibitor: as a solvation scoring function
- Identify the key pharmacohore feature
- Halogen bond interaction design
1. Load the script from flare python gui: Flare | Python | Python Interpretor |Load .. (Figure 1. step 1, 2 and 3) gist_dG_at_picked_atom_flare_python_gui.py
2. select the protein intereted (Figure 1. step 4).
3. pick a atom, single click button Run (Figure 1. step 5 and 6).
Figure 1. Five steps to use the script
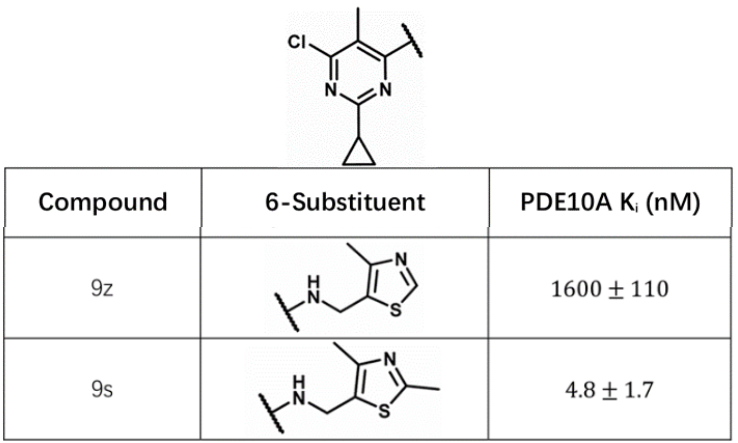
This section will introduce how to calculate the ΔGwatdisp for ligands 9s and 9z.
Enter the following command in the shell:
gist_dG_watdisp.py -g 5c29_apo_gist_dG.dx -i 9s-9z.sdf -o 9s-9z_out.sdfYou can find the GIST-dG-Watdisp tag in the output SDF file, which represents the calculated ΔGwatdisp. You can find the calculation results for 9s as follows:
> <GIST-dG-Watdisp> (1)
-41.599And ΔGwatdisp for 9z as follows:
> <GIST-dG-Watdisp> (2)
-38.223Thus, the ΔΔGwatdisp between 9s and 9z is 3.38 kcal/mol, which is very close to the difference in their binding free energies (ΔΔG = 3.44 kcal/mol).
rdkit, numpy and GridDataFormats are required.
- Yoshida, S.; Uehara, S.; Kondo, N.; Takahashi, Y.; Yamamoto, S.; Kameda, A.; Kawagoe, S.; Inoue, N.; Yamada, M.; Yoshimura, N.; et al. Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides. 2022. https://doi.org/10.1021/acs.jmedchem.2c00919.
- Yang, Z.; Xiaoyun, L. U. The Role of Water Molecules in Drug Design. Prog. Pharm. Sci. 2022, 46 (1), 47–59. https://pps.cpu.edu.cn/article/id/ba95392b-7a5e-44d4-a2b5-a3084711d6f5
- Targowska-Duda, K. M.; Maj, M.; Drączkowski, P.; Budzyńska, B.; Boguszewska-Czubara, A.; Wróbel, T. M.; Laitinen, T.; Kaczmar, P.; Poso, A.; Kaczor, A. A. WaterMap Guided Structure‐based Virtual Screening for Acetylcholinesterase Inhibitors. ChemMedChem 2022, n/a (n/a). https://doi.org/10.1002/cmdc.202100721.
- Zsidó, B. Z.; Hetényi, C. The Role of Water in Ligand Binding. Curr. Opin. Struct. Biol. 2021, 67 (Figure 1), 1–8. https://doi.org/10.1016/j.sbi.2020.08.002.
- Lloyd, M. G.; Huckvale, R.; Cheung, K. J.; Rodrigues, M. J.; Collie, G. W.; Pierrat, O. A.; Gatti Iou, M.; Carter, M.; Davis, O. A.; McAndrew, P. C.; et al. Into Deep Water: Optimizing BCL6 Inhibitors by Growing into a Solvated Pocket. J. Med. Chem. 2021, 64 (23), 17079–17097. https://doi.org/10.1021/acs.jmedchem.1c00946.
- Hüfner-Wulsdorf, T.; Klebe, G. Mapping Water Thermodynamics on Drug Candidates via Molecular Building Blocks: A Strategy to Improve Ligand Design and Rationalize SAR. J. Med. Chem. 2021, 64 (8), 4662–4676. https://doi.org/10.1021/acs.jmedchem.0c02115.
- Andreev, S.; Pantsar, T.; Tesch, R.; Kahlke, N.; El-Gokha, A.; Ansideri, F.; Grätz, L.; Romasco, J.; Sita, G.; Geibel, C.; et al. Addressing a Trapped High-Energy Water: Design and Synthesis of Highly Potent Pyrimidoindole-Based Glycogen Synthase Kinase-3β Inhibitors. J. Med. Chem. 2021, acs.jmedchem.0c02146. https://doi.org/10.1021/acs.jmedchem.0c02146.
- Hüfner-Wulsdorf, T.; Klebe, G. Protein–Ligand Complex Solvation Thermodynamics: Development, Parameterization, and Testing of GIST-Based Solvent Functionals. J. Chem. Inf. Model. 2020, 60 (3), 1409–1423. https://doi.org/10.1021/acs.jcim.9b01109.
- Yoshidome, T.; Ikeguchi, M.; Ohta, M. Comprehensive 3D-RISM Analysis of the Hydration of Small Molecule Binding Sites in Ligand-Free Protein Structures. J. Comput. Chem. 2020, 41 (28), 2406–2419. https://doi.org/10.1002/jcc.26406.
- Hüfner-Wulsdorf, T.; Klebe, G. Advancing GIST-Based Solvent Functionals through Multiobjective Optimization of Solvent Enthalpy and Entropy Scoring Terms. J. Chem. Inf. Model. 2020, 60 (12), 6654–6665. https://doi.org/10.1021/acs.jcim.0c01133.
- Li, Y.; Gao, Y.; Holloway, M. K.; Wang, R. Prediction of the Favorable Hydration Sites in a Protein Binding Pocket and Its Application to Scoring Function Formulation. J. Chem. Inf. Model. 2020, 60 (9), 4359–4375. https://doi.org/10.1021/acs.jcim.9b00619.
- Viviani, L. G.; Piccirillo, E.; Ulrich, H.; Amaral, A. T. -d. T. D. Virtual Screening Approach for the Identification of Hydroxamic Acids as Novel Human Ecto-5′-Nucleotidase Inhibitors. J. Chem. Inf. Model. 2020, 60 (2), 621–630. https://doi.org/10.1021/acs.jcim.9b00884.
- Bancet, A.; Raingeval, C.; Lomberget, T.; Le Borgne, M.; Guichou, J.-F.; Krimm, I. Fragment Linking Strategies for Structure-Based Drug Design. J. Med. Chem. 2020, 63 (20), 11420–11435. https://doi.org/10.1021/acs.jmedchem.0c00242.
- Gerstenberger, B. S.; Ambler, C.; Arnold, E. P.; Banker, M.; Brown, M. F.; Clark, J. D.; Dermenci, A.; Dowty, M. E.; Fensome, A.; Fish, S.; et al. Discovery of Tyrosine Kinase 2 (TYK2) Inhibitor (PF-06826647) for the Treatment of Autoimmune Diseases. J. Med. Chem. 2020, 63 (22), 13561–13577. https://doi.org/10.1021/acs.jmedchem.0c00948.
- Wang, Y.; Fu, Q.; Zhou, Y.; Du, Y.; Huang, N. Replacement of Protein Binding-Site Waters Contributes to Favorable Halogen Bond Interactions. J. Chem. Inf. Model. 2019, 59 (7), 3136–3143. https://doi.org/10.1021/acs.jcim.9b00128.
- Schaller, D.; Pach, S.; Wolber, G. PyRod: Tracing Water Molecules in Molecular Dynamics Simulations. J. Chem. Inf. Model. 2019, No. May, acs.jcim.9b00281. https://doi.org/10.1021/acs.jcim.9b00281.
- Nittinger, E.; Gibbons, P.; Eigenbrot, C.; Davies, D. R.; Maurer, B.; Yu, C. L.; Kiefer, J. R.; Kuglstatter, A.; Murray, J.; Ortwine, D. F.; et al. Water Molecules in Protein–Ligand Interfaces. Evaluation of Software Tools and SAR Comparison. J. Comput. Aided. Mol. Des. 2019, 33 (3), 307–330. https://doi.org/10.1007/s10822-019-00187-y.
- Nguyen, C.; Yamazaki, T.; Kovalenko, A.; Case, D. A.; Gilson, M. K.; Kurtzman, T.; Luchko, T. A Molecular Reconstruction Approach to Site-Based 3D-RISM and Comparison to GIST Hydration Thermodynamic Maps in an Enzyme Active Site. PLoS One 2019, 14 (7), e0219473. https://doi.org/10.1371/journal.pone.0219473.
- Lu, J.; Hou, X.; Wang, C.; Zhang, Y. Incorporating Explicit Water Molecules and Ligand Conformation Stability in Machine-Learning Scoring Functions. J. Chem. Inf. Model. 2019, 59 (11), 4540–4549. https://doi.org/10.1021/acs.jcim.9b00645.
- Bucher, D.; Stouten, P.; Triballeau, N. Shedding Light on Important Waters for Drug Design: Simulations versus Grid-Based Methods. J. Chem. Inf. Model. 2018, 58 (3), 692–699. https://doi.org/10.1021/acs.jcim.7b00642.
- Nittinger, E.; Flachsenberg, F.; Bietz, S.; Lange, G.; Klein, R.; Rarey, M. Placement of Water Molecules in Protein Structures: From Large-Scale Evaluations to Single-Case Examples. J. Chem. Inf. Model. 2018, 58 (8), 1625–1637. https://doi.org/10.1021/acs.jcim.8b00271.
- Hamaguchi, H.; Amano, Y.; Moritomo, A.; Shirakami, S.; Nakajima, Y.; Nakai, K.; Nomura, N.; Ito, M.; Higashi, Y.; Inoue, T. Discovery and Structural Characterization of Peficitinib (ASP015K) as a Novel and Potent JAK Inhibitor. Bioorg. Med. Chem. 2018, 26 (18), 4971–4983. https://doi.org/10.1016/j.bmc.2018.08.005.
- Wang, Y.; Du, Y.; Huang, N. A Survey of the Role of Nitrile Groups in Protein–Ligand Interactions. Future Med. Chem. 2018, 10 (23), 2713–2728. https://doi.org/10.4155/fmc-2018-0252.
- Masters, M. R.; Mahmoud, A. H.; Yang, Y.; Lill, M. A. Efficient and Accurate Hydration Site Profiling for Enclosed Binding Sites. J. Chem. Inf. Model. 2018, 58 (11), 2183–2188. https://doi.org/10.1021/acs.jcim.8b00544.
- Ahmad, S.; Shaker, B.; Ahmad, F.; Raza, S.; Azam, S. S. Moleculer Dynamics Simulaiton Revealed Reciever Domain of Acinetobacter Baumannii BfmR Enzyme as the Hot Spot for Future Antibiotics Designing. J. Biomol. Struct. Dyn. 2018, 1–42. https://doi.org/10.1080/07391102.2018.1498805.
- Ahmad, S.; Raza, S.; Abro, A.; Liedl, K. R.; Azam, S. S. Toward Novel Inhibitors against KdsB: A Highly Specific and Selective Broad-Spectrum Bacterial Enzyme. J. Biomol. Struct. Dyn. 2018, 1–20. https://doi.org/10.1080/07391102.2018.1459318.
- Matter, H.; Güssregen, S. Characterizing Hydration Sites in Protein-Ligand Complexes towards the Design of Novel Ligands. Bioorg. Med. Chem. Lett. 2018, 28 (14), 2343–2352. https://doi.org/10.1016/j.bmcl.2018.05.061.
- Callegari, D.; Ranaghan, K. E.; Woods, C. J.; Minari, R.; Tiseo, M.; Mor, M.; Mulholland, A. J.; Lodola, A. L718Q Mutant EGFR Escapes Covalent Inhibition by Stabilizing a Non-Reactive Conformation of the Lung Cancer Drug Osimertinib. Chem. Sci. 2018, 9 (10), 2740–2749. https://doi.org/10.1039/C7SC04761D.
- Cappel, D.; Sherman, W.; Beuming, T. Calculating Water Thermodynamics in the Binding Site of Proteins – Applications of WaterMap to Drug Discovery. Curr. Top. Med. Chem. 2017, 17 (23), 2586–2598. https://doi.org/10.2174/1568026617666170414141452.
- Spyrakis, F.; Ahmed, M. H.; Bayden, A. S.; Cozzini, P.; Mozzarelli, A.; Kellogg, G. E. The Roles of Water in the Protein Matrix: A Largely Untapped Resource for Drug Discovery. J. Med. Chem. 2017, 60 (16), 6781–6828. https://doi.org/10.1021/acs.jmedchem.7b00057.
- Schauperl, M.; Czodrowski, P.; Fuchs, J. E.; Huber, R. G.; Waldner, B. J.; Podewitz, M.; Kramer, C.; Liedl, K. R. Binding Pose Flip Explained via Enthalpic and Entropic Contributions. J. Chem. Inf. Model. 2017, 57 (2), 345–354. https://doi.org/10.1021/acs.jcim.6b00483.
- Balius, T. E.; Fischer, M.; Stein, R. M.; Adler, T. B.; Nguyen, C. N.; Cruz, A.; Gilson, M. K.; Kurtzman, T.; Shoichet, B. K. Testing Inhomogeneous Solvation Theory in Structure-Based Ligand Discovery. Proc. Natl. Acad. Sci. 2017, 114 (33), E6839–E6846. https://doi.org/10.1073/pnas.1703287114.
- Güssregen, S.; Matter, H.; Hessler, G.; Lionta, E.; Heil, J.; Kast, S. M. Thermodynamic Characterization of Hydration Sites from Integral Equation-Derived Free Energy Densities: Application to Protein Binding Sites and Ligand Series. J. Chem. Inf. Model. 2017, 57 (7), 1652–1666. https://doi.org/10.1021/acs.jcim.6b00765.
- Czodrowski, P.; Mallinger, A.; Wienke, D.; Esdar, C.; Pöschke, O.; Busch, M.; Rohdich, F.; Eccles, S. A.; Ortiz-Ruiz, M.-J.; Schneider, R.; et al. Structure-Based Optimization of Potent, Selective, and Orally Bioavailable CDK8 Inhibitors Discovered by High-Throughput Screening. J. Med. Chem. 2016, 59 (20), 9337–9349. https://doi.org/10.1021/acs.jmedchem.6b00597.
- Bodnarchuk, M. S. Water, Water, Everywhere... It’s Time to Stop and Think. Drug Discov. Today 2016, 21 (7), 1139–1146. https://doi.org/10.1016/j.drudis.2016.05.009.
- Myrianthopoulos, V.; Gaboriaud-Kolar, N.; Tallant, C.; Hall, M.-L.; Grigoriou, S.; Brownlee, P. M.; Fedorov, O.; Rogers, C.; Heidenreich, D.; Wanior, M.; et al. Discovery and Optimization of a Selective Ligand for the Switch/Sucrose Nonfermenting-Related Bromodomains of Polybromo Protein-1 by the Use of Virtual Screening and Hydration Analysis. J. Med. Chem. 2016, 59 (19), 8787–8803. https://doi.org/10.1021/acs.jmedchem.6b00355.
- Ramsey, S.; Nguyen, C.; Salomon-Ferrer, R.; Walker, R. C.; Gilson, M. K.; Kurtzman, T. Solvation Thermodynamic Mapping of Molecular Surfaces in AmberTools: GIST. J. Comput. Chem. 2016, 37 (21), 2029–2037. https://doi.org/10.1002/jcc.24417.
- Calabrò, G.; Woods, C. J.; Powlesland, F.; Mey, A. S. J. S.; Mulholland, A. J.; Michel, J. Elucidation of Nonadditive Effects in Protein–Ligand Binding Energies: Thrombin as a Case Study. J. Phys. Chem. B 2016, 120 (24), 5340–5350. https://doi.org/10.1021/acs.jpcb.6b03296.
- Zoidis, G.; Giannakopoulou, E.; Stevaert, A.; Frakolaki, E.; Myrianthopoulos, V.; Fytas, G.; Mavromara, P.; Mikros, E.; Bartenschlager, R.; Vassilaki, N.; et al. Novel Indole–Flutimide Heterocycles with Activity against Influenza PA Endonuclease and Hepatitis C Virus. Medchemcomm 2016, 7 (3), 447–456. https://doi.org/10.1039/C5MD00439J.
- Murphy, R. B.; Repasky, M. P.; Greenwood, J. R.; Tubert-Brohman, I.; Jerome, S.; Annabhimoju, R.; Boyles, N. A.; Schmitz, C. D.; Abel, R.; Farid, R.; et al. WScore: A Flexible and Accurate Treatment of Explicit Water Molecules in Ligand-Receptor Docking. J. Med. Chem. 2016, 59 (9), 4364–4384. https://doi.org/10.1021/acs.jmedchem.6b00131.
- Uehara, S.; Tanaka, S. AutoDock-GIST: Incorporating Thermodynamics of Active-Site Water into Scoring Function for Accurate Protein-Ligand Docking. Molecules 2016, 21 (11), 1604. https://doi.org/10.3390/molecules21111604.
- Horbert, R.; Pinchuk, B.; Johannes, E.; Schlosser, J.; Schmidt, D.; Cappel, D.; Totzke, F.; Schächtele, C.; Peifer, C. Optimization of Potent Dfg-in Inhibitors of Platelet Derived Growth Factor Receptorβ (PDGF-Rβ) Guided by Water Thermodynamics. J. Med. Chem. 2015, 58 (1), 170–182. https://doi.org/10.1021/jm500373x.
- Bayden, A. S.; Moustakas, D. T.; Joseph-McCarthy, D.; Lamb, M. L. Evaluating Free Energies of Binding and Conservation of Crystallographic Waters Using SZMAP. J. Chem. Inf. Model. 2015, 55 (8), 1552–1565. https://doi.org/10.1021/ci500746d.
- Nguyen, C. N.; Cruz, A.; Gilson, M. K.; Kurtzman, T. Thermodynamics of Water in an Enzyme Active Site: Grid-Based Hydration Analysis of Coagulation Factor Xa. chemrxiv 2014, 10 (7), 2769–2780. https://doi.org/10.1021/ct401110x.
- Czodrowski, P.; Hölzemann, G.; Barnickel, G.; Greiner, H.; Musil, D. Selection of Fragments for Kinase Inhibitor Design: Decoration Is Key. J. Med. Chem. 2015, 58 (1), 457–465. https://doi.org/10.1021/jm501597j.
- Smith, C. R.; Dougan, D. R.; Komandla, M.; Kanouni, T.; Knight, B.; Lawson, J. D.; Sabat, M.; Taylor, E. R.; Vu, P.; Wyrick, C. Fragment-Based Discovery of a Small Molecule Inhibitor of Bruton’s Tyrosine Kinase. J. Med. Chem. 2015, 58 (14), 5437–5444. https://doi.org/10.1021/acs.jmedchem.5b00734.
- Bortolato, A.; Tehan, B. G.; Bodnarchuk, M. S.; Essex, J. W.; Mason, J. S. Water Network Perturbation in Ligand Binding: Adenosine A 2A Antagonists as a Case Study. J. Chem. Inf. Model. 2013, 53 (7), 1700–1713. https://doi.org/10.1021/ci4001458.
- Breiten, B.; Lockett, M. R.; Sherman, W.; Fujita, S.; Al-Sayah, M.; Lange, H.; Bowers, C. M.; Heroux, A.; Krilov, G.; Whitesides, G. M. Water Networks Contribute to Enthalpy/Entropy Compensation in Protein–Ligand Binding. J. Am. Chem. Soc. 2013, 135 (41), 15579–15584. https://doi.org/10.1021/ja4075776.
- Woods, C. J.; Malaisree, M.; Long, B.; McIntosh-Smith, S.; Mulholland, A. J. Computational Assay of H7n9 Influenza Neuraminidase Reveals R292k Mutation Reduces Drug Binding Affinity. Sci. Rep. 2013, 3, 7–12. https://doi.org/10.1038/srep03561.
- Kohlmann, A.; Zhu, X.; Dalgarno, D. Application of MM-GB/SA and WaterMap to SRC Kinase Inhibitor Potency Prediction. ACS Med. Chem. Lett. 2012, 3 (2), 94–99. https://doi.org/10.1021/ml200222u.
- Beuming, T.; Che, Y.; Abel, R.; Kim, B.; Shanmugasundaram, V.; Sherman, W. Thermodynamic Analysis of Water Molecules at the Surface of Proteins and Applications to Binding Site Prediction and Characterization. Proteins Struct. Funct. Bioinforma. 2012, 80 (3), 871–883. https://doi.org/10.1002/prot.23244.
- Trujillo, J. I.; Kiefer, J. R.; Huang, W.; Day, J. E.; Moon, J.; Jerome, G. M.; Bono, C. P.; Kornmeier, C. M.; Williams, M. L.; Kuhn, C.; et al. Investigation of the Binding Pocket of Human Hematopoietic Prostaglandin (PG) D2 Synthase (HH-PGDS): A Tale of Two Waters. Bioorg. Med. Chem. Lett. 2012, 22 (11), 3795–3799. https://doi.org/10.1016/j.bmcl.2012.04.004.
- Mason, J. S.; Bortolato, A.; Congreve, M.; Marshall, F. H. New Insights from Structural Biology into the Druggability of G Protein-Coupled Receptors. Trends Pharmacol. Sci. 2012, 33 (5), 249–260. https://doi.org/10.1016/j.tips.2012.02.005.
- Kung, P.-P.; Sinnema, P.-J.; Richardson, P.; Hickey, M. J.; Gajiwala, K. S.; Wang, F.; Huang, B.; McClellan, G.; Wang, J.; Maegley, K.; et al. Design Strategies to Target Crystallographic Waters Applied to the Hsp90 Molecular Chaperone. Bioorg. Med. Chem. Lett. 2011, 21 (12), 3557–3562. https://doi.org/10.1016/j.bmcl.2011.04.130.
- Nguyen, C.; Gilson, M. K.; Young, T. Structure and Thermodynamics of Molecular Hydration via Grid Inhomogeneous Solvation Theory. ChemRxiv 2011.
- Robinson, D. D.; Sherman, W.; Farid, R. Understanding Kinase Selectivity Through Energetic Analysis of Binding Site Waters. ChemMedChem 2010, 5 (4), 618–627. https://doi.org/10.1002/cmdc.200900501.
- Michel, J.; Tirado-Rives, J.; Jorgensen, W. L. Energetics of Displacing Water Molecules from Protein Binding Sites: Consequences for Ligand Optimization. J. Am. Chem. Soc. 2009, 131 (42), 15403–15411. https://doi.org/10.1021/ja906058w.
- Abel, R.; Young, T.; Farid, R.; Berne, B. J.; Friesner, R. A. Role of the Active-Site Solvent in the Thermodynamics of Factor Xa Ligand Binding. J. Am. Chem. Soc. 2008, 130 (9), 2817–2831. https://doi.org/10.1021/ja0771033.
- Young, T.; Abel, R.; Kim, B.; Berne, B. J.; Friesner, R. A. Motifs for Molecular Recognition Exploiting Hydrophobic Enclosure in Protein–Ligand Binding. Proc. Natl. Acad. Sci. 2007, 104 (3), 808–813. https://doi.org/10.1073/pnas.0610202104.
- Chen, J. M.; Xu, S. L.; Wawrzak, Z.; Basarab, G. S.; Jordan, D. B. Structure-Based Design of Potent Inhibitors of Scytalone Dehydratase: Displacement of a Water Molecule from the Active Site. Biochemistry 1998, 37 (51), 17735–17744. https://doi.org/10.1021/bi981848r.