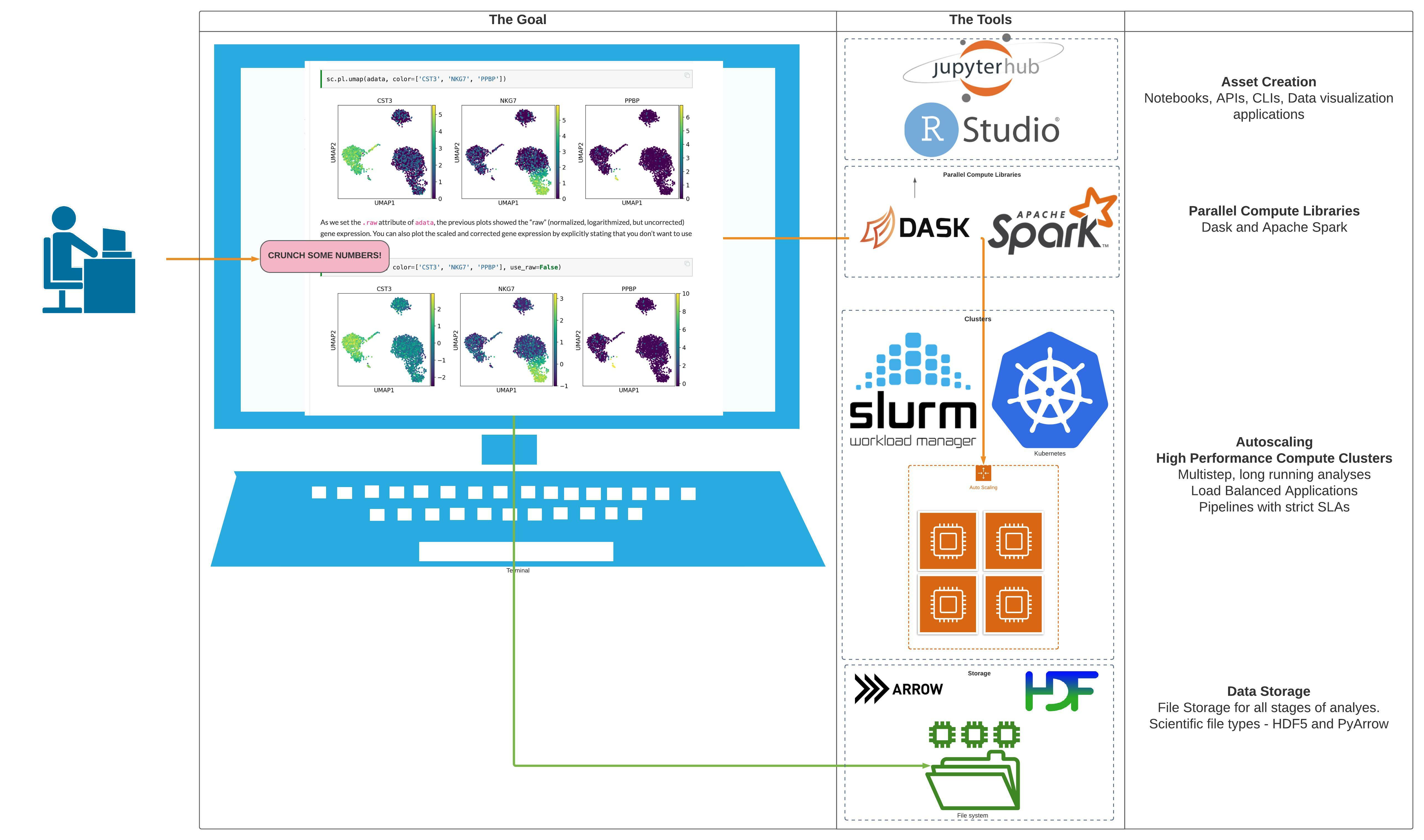
The terraform-aws-batch recipe provides a starter template for getting started with AWS Batch. It creates:
- AWS Batch Compute Environment ** Configured for use with the AWS Secrets Manager
- AWS Job Queue associated to the created Compute Environment
This project is part of the "BioAnalyze" project, which aims to make High Performance Compute Architecture accessible to everyone.
It's 100% Open Source and licensed under the APACHE2.
IMPORTANT: We do not pin modules to versions in our examples because of the difficulty of keeping the versions in the documentation in sync with the latest released versions. We highly recommend that in your code you pin the version to the exact version you are using so that your infrastructure remains stable, and update versions in a systematic way so that they do not catch you by surprise.
Also, because of a bug in the Terraform registry (hashicorp/terraform#21417), the registry shows many of our inputs as required when in fact they are optional. The table below correctly indicates which inputs are required.
For a complete example, see examples/complete.
Each of the examples are deployed using GitHub actions. For more information see the examples and the .github directories.
More complete documentation and tutorials coming soon!
The examples for using this module are in the examples directory.
Available targets:
help Help screen
help/all Display help for all targets
help/short This help short screen
lint Lint terraform code
| Name | Version |
|---|---|
| terraform | >= 0.13 |
| local | >= 1.2 |
| random | >= 2.2 |
| Name | Version |
|---|---|
| aws | 3.49.0 |
| Name | Source | Version |
|---|---|---|
| ec2_batch_compute_environment | ./modules/aws-batch-ec2 | n/a |
| fargate_batch_compute_environment | ./modules/aws-batch-fargate | n/a |
| this | cloudposse/label/null | 0.24.1 |
| Name | Type |
|---|---|
| aws_batch_job_queue.default_queue | resource |
| aws_default_security_group.default | resource |
| aws_iam_instance_profile.ecs_instance_role | resource |
| aws_iam_policy.secrets_full_access | resource |
| aws_iam_role.aws_batch_service_role | resource |
| aws_iam_role.batch_execution_role | resource |
| aws_iam_role.ecs_instance_role | resource |
| aws_iam_role_policy_attachment.aws_batch_full_access | resource |
| aws_iam_role_policy_attachment.aws_batch_service_role | resource |
| aws_iam_role_policy_attachment.batch_execution_attach_secrets | resource |
| aws_iam_role_policy_attachment.batch_execution_role | resource |
| aws_iam_role_policy_attachment.ecs_instance_role | resource |
| aws_secretsmanager_secret.batch | resource |
| aws_security_group.batch | resource |
| aws_caller_identity.current | data source |
| aws_iam_policy_document.secrets_full_access | data source |
| aws_vpc.selected | data source |
| Name | Description | Type | Default | Required |
|---|---|---|---|---|
| additional_tag_map | Additional tags for appending to tags_as_list_of_maps. Not added to tags. |
map(string) |
{} |
no |
| additional_user_data | Additional User Data for the launch template. Must include ==MYBOUNDARY== and Content-Type: entries. | string |
"" |
no |
| ami_owners | List of owners for source ECS AMI. | list(any) |
[ |
no |
| attributes | Additional attributes (e.g. 1) |
list(string) |
[] |
no |
| bid_percentage | Integer of minimum percentage that a Spot Instance price must be when compared to on demand. Example: A value of 20 would require the spot price be lower than 20% the current on demand price. | string |
"100" |
no |
| block_device_mappings | Specify volumes to attach to the instance besides the volumes specified by the AMI | list(object({ |
[] |
no |
| context | Single object for setting entire context at once. See description of individual variables for details. Leave string and numeric variables as null to use default value.Individual variable settings (non-null) override settings in context object, except for attributes, tags, and additional_tag_map, which are merged. |
any |
{ |
no |
| credit_specification | Customize the credit specification of the instances | object({ |
null |
no |
| custom_ami | Optional string for custom AMI. If omitted, latest ECS AMI in the current region will be used. | string |
"" |
no |
| delimiter | Delimiter to be used between namespace, environment, stage, name and attributes.Defaults to - (hyphen). Set to "" to use no delimiter at all. |
string |
null |
no |
| disable_api_termination | If true, enables EC2 Instance Termination Protection |
bool |
false |
no |
| docker_max_container_size | If docker_expand_volume is true, containers will allocate this amount of storage (GB) when launched. | number |
50 |
no |
| ebs_optimized | If true, the launched EC2 instance will be EBS-optimized | bool |
false |
no |
| ec2_key_pair | Optional keypair to connect to the instance with. Consider SSM as an alternative. | string |
"" |
no |
| elastic_gpu_specifications | Specifications of Elastic GPU to attach to the instances | object({ |
null |
no |
| enable_monitoring | Enable/disable detailed monitoring | bool |
true |
no |
| enabled | Set to false to prevent the module from creating any resources | bool |
null |
no |
| environment | Environment, e.g. 'uw2', 'us-west-2', OR 'prod', 'staging', 'dev', 'UAT' | string |
null |
no |
| iam_instance_profile_name | The IAM instance profile name to associate with launched instances | string |
"" |
no |
| id_length_limit | Limit id to this many characters (minimum 6).Set to 0 for unlimited length.Set to null for default, which is 0.Does not affect id_full. |
number |
null |
no |
| instance_initiated_shutdown_behavior | Shutdown behavior for the instances. Can be stop or terminate |
string |
"terminate" |
no |
| instance_market_options | The market (purchasing) option for the instances | object({ |
null |
no |
| instance_types | Optional list of instance types. | list(any) |
[ |
no |
| key_name | The SSH key name that should be used for the instance | string |
"" |
no |
| label_key_case | The letter case of label keys (tag names) (i.e. name, namespace, environment, stage, attributes) to use in tags.Possible values: lower, title, upper.Default value: title. |
string |
null |
no |
| label_order | The naming order of the id output and Name tag. Defaults to ["namespace", "environment", "stage", "name", "attributes"]. You can omit any of the 5 elements, but at least one must be present. |
list(string) |
null |
no |
| label_value_case | The letter case of output label values (also used in tags and id).Possible values: lower, title, upper and none (no transformation).Default value: lower. |
string |
null |
no |
| max_vcpus | Max vCPUs. Default 2 for m4.large. | string |
8 |
no |
| metadata_http_endpoint_enabled | Set false to disable the Instance Metadata Service. | bool |
true |
no |
| metadata_http_put_response_hop_limit | The desired HTTP PUT response hop limit (between 1 and 64) for Instance Metadata Service requests. The default is 2 to support containerized workloads. |
number |
2 |
no |
| metadata_http_tokens_required | Set true to require IMDS session tokens, disabling Instance Metadata Service Version 1. | bool |
true |
no |
| min_vcpus | Minimum vCPUs. > 0 causes instances to always be running. | string |
0 |
no |
| name | Solution name, e.g. 'app' or 'jenkins' | string |
null |
no |
| namespace | Namespace, which could be your organization name or abbreviation, e.g. 'eg' or 'cp' | string |
null |
no |
| placement | The placement specifications of the instances | object({ |
null |
no |
| regex_replace_chars | Regex to replace chars with empty string in namespace, environment, stage and name.If not set, "/[^a-zA-Z0-9-]/" is used to remove all characters other than hyphens, letters and digits. |
string |
null |
no |
| region | AWS Region | string |
"us-east-1" |
no |
| secrets_enabled | Enable IAM Role for AWS Secrets Manager | bool |
false |
no |
| security_group_ids | List of additional security groups to associate with cluster instances. If empty, default security group will be added. | list(any) |
[ |
no |
| stage | Stage, e.g. 'prod', 'staging', 'dev', OR 'source', 'build', 'test', 'deploy', 'release' | string |
null |
no |
| subnet_ids | List of subnets compute environment instances will be deployed in. | list(string) |
n/a | yes |
| tag_specifications_resource_types | List of tag specification resource types to tag. Valid values are instance, volume, elastic-gpu and spot-instances-request. | set(string) |
[ |
no |
| tags | Additional tags (e.g. map('BusinessUnit','XYZ') |
map(string) |
{} |
no |
| type | AWS Batch Compute Environment Type: must be one of EC2, SPOT, FARGATE or FARGATE_SPOT. | string |
"EC2" |
no |
| vpc_id | VPC ID | string |
n/a | yes |
| Name | Description |
|---|---|
| account_id | n/a |
| aws_batch_compute_environment | n/a |
| aws_batch_ecs_instance_role | n/a |
| aws_batch_execution_role | n/a |
| aws_batch_job_queue | n/a |
| aws_batch_service_role | n/a |
| aws_iam_policy_document-secrets_full_access | n/a |
| aws_secrets_manager_secret-batch | n/a |
| caller_arn | n/a |
| caller_user | n/a |
| id | ID of the created example |
Like this project? Please give it a ★ on our GitHub! (it helps a lot)
Check out these related projects.
- terraform-aws-eks-autoscaling - Wrapper module for terraform-aws-eks-cluster, terraform-aws-eks-worker, and terraform-aws-eks-node-group
- terraform-aws-eks-cluster - Base CloudPosse module for AWS EKS Clusters"
- terraform-null-label - Terraform module designed to generate consistent names and tags for resources. Use terraform-null-label to implement a strict naming convention.
For additional context, refer to some of these links.
- Terraform Standard Module Structure - HashiCorp's standard module structure is a file and directory layout we recommend for reusable modules distributed in separate repositories.
- Terraform Module Requirements - HashiCorp's guidance on all the requirements for publishing a module. Meeting the requirements for publishing a module is extremely easy.
- Terraform
batch_compute_environmentResource - Creates a AWS Batch compute environment. Compute environments contain the Amazon ECS container instances that are used to run containerized batch jobs. - Terraform
batch_job_queueResource - Provides a Batch Job Queue resource. - Terraform `batch_job_definition - Provides a Batch Job Definition resource.
- Terraform
random_integerResource - The resource random_integer generates random values from a given range, described by the min and max attributes of a given resource. - Terraform Version Pinning - The required_version setting can be used to constrain which versions of the Terraform CLI can be used with your configuration
Got a question? We got answers.
File a GitHub issue, send us an jillian@dabbleofdevops.com.
I'll help you build your data science cloud infrastructure from the ground up so you can own it using open source software. Then I'll show you how to operate it and stick around for as long as you need us.
Work directly with me via email, slack, and video conferencing.
- Scientific Workflow Automation and Optimization. Got workflows that are giving you trouble? Let's work together to ensure that your analyses run with or without your scientists being fully caffeinated.
- High Performance Compute Infrastructure. Highly available, auto scaling clusters to analyze all the (bioinformatics related!) things. All setups are completely integrated with your workflow system of choice, whether that is Airflow, Prefect, Snakemake or Nextflow.
- Kubernetes and AWS Batch Setup for Apache Airflow Orchestrate your Bioinformatics Workflows with Apache Airflow. Get full auditing, SLA, logging and monitoring for your workflows running on AWS Batch.
- High Performance Compute Setup that Int You'll have built-in governance with accountability and audit logs for all changes.
- Docker Images Get advice and hands on training for your team to build complex software stacks onto docker images.
- Training. You'll receive hands-on training so your team can operate what we build.
- Questions. You'll have a direct line of communication between our teams via a Shared Slack channel.
- Troubleshooting. You'll get help to triage when things aren't working.
- Bug Fixes. We'll rapidly work with you to fix any bugs in our projects.
Please use the issue tracker to report any bugs or file feature requests.
If you are interested in being a contributor and want to get involved in developing this project or help out with other projects, I would love to hear from you! Shoot me an email at jillian@dabbleofdevops.com.
In general, PRs are welcome. We follow the typical "fork-and-pull" Git workflow.
- Fork the repo on GitHub
- Clone the project to your own machine
- Commit changes to your own branch
- Push your work back up to your fork
- Submit a Pull Request so that we can review your changes
The README.md is created using the standard CloudPosse template that has been modified to use BioAnalyze information and URLs, and other documentation is generated using jupyter-book.
Terraform code does not render properly when using the literalinclude directive, so instead we use pygmentize to render it to html which is included directly.
.. raw:: html
:file: ./_html/main.tf.html
NOTE: Be sure to merge the latest changes from "upstream" before making a pull request!
Copyright © 2021-2021 Dabble of DevOps, SCorp
See LICENSE for full details.
Licensed to the Apache Software Foundation (ASF) under one
or more contributor license agreements. See the NOTICE file
distributed with this work for additional information
regarding copyright ownership. The ASF licenses this file
to you under the Apache License, Version 2.0 (the
"License"); you may not use this file except in compliance
with the License. You may obtain a copy of the License at
https://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing,
software distributed under the License is distributed on an
"AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY
KIND, either express or implied. See the License for the
specific language governing permissions and limitations
under the License.
All other trademarks referenced herein are the property of their respective owners.
|
Jillian Rowe |
Learn more at Dabble of DevOps