-
Notifications
You must be signed in to change notification settings - Fork 0
shapeevolution iterative
This log file refers to the code
script_iterevolution.m.
One can see in the larger frame leaps jx>17 that the segmentation is
qualitatively worse. Originally, the focus of this approach is to update
the shape of the unknown frame at each point and take that information onto
the next frame.
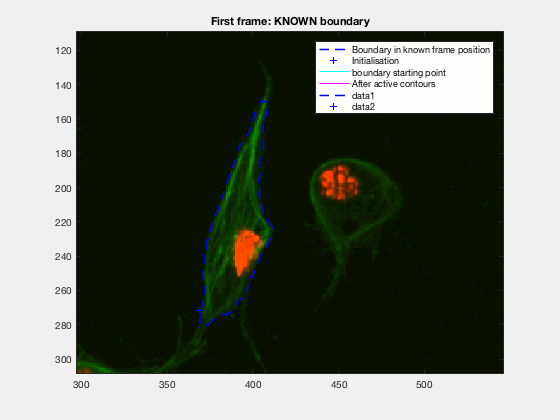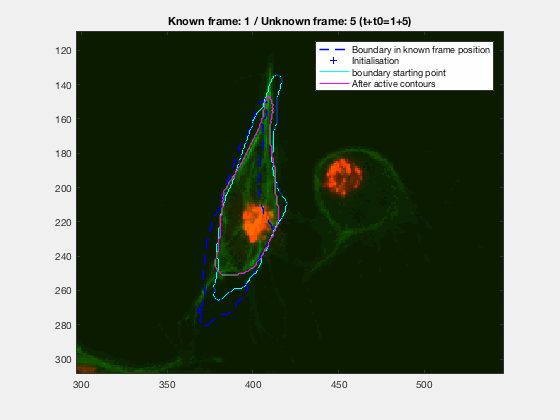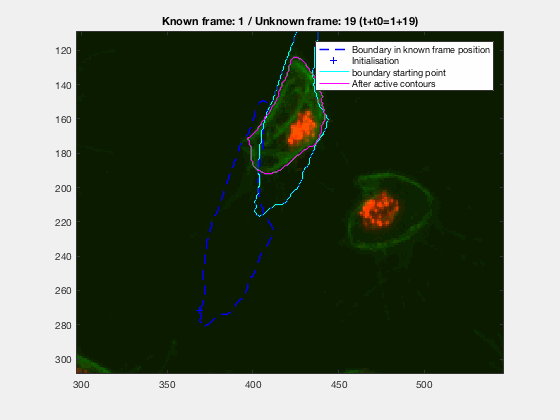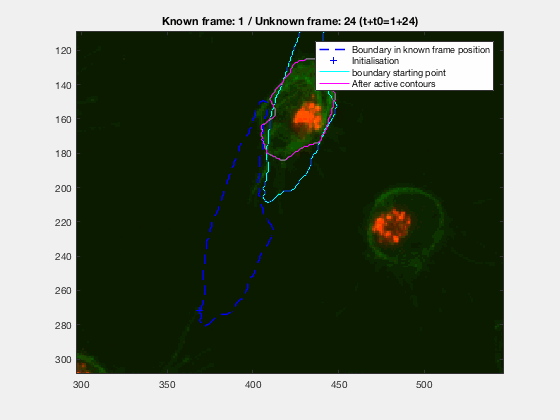
This initial test show promise, however, in this case, the known, knownfr and
unknown ukfr were chosen based on leaps tj = t0+jx. A test trying to
construct the frames at each time frame is necessary, in a way that only
the information from the previous frame is carried out to the next one,
this means: tj = t(j-1) + 1, t(j-1)=t(j-2)+1, ..., t2 = t1+1, t1=t0+1;
where the ONLY completely known frame will be t0, and the rest will
be constructed along the way.
The script initscript.m is called, loading the relevant
variables to the workspace. Then, a clump is chosen. The function
searchClumpOnFrame.m can be used to identify
a clump to load. The usual clumps tested are, the ones where the frames that
span the life of the clump are specified:
-
wuc=8002useful tracks:trackinfo(trackinfo.timeframe<418,:) = []; -
wuc=8007useful tracks:trackinfo(~ismember(trackinfo.timeframe, 15:50),:)=[]; -
wuc=8007005useful tracks:trackinfo(~ismember(trackinfo.timeframe, 15:18),:)=[]; -
wuc=11010useful tracks:trackinfo(trackinfo.timeframe>=144,:)=[]; -
wuc=14013, useful trackstrackinfo(~ismember(trackinfo.timeframe, 376:386),:)=[]; wuc=60010wuc=60010002wuc=15014013
% for clump 8002
trackinfo(trackinfo.timeframe<418,:) = [];
% ORRRR, ....
trackinfo(~ismember(trackinfo.timeframe, 418:495),:)=[];
%
% for clump 11010
trackinfo(trackinfo.timeframe>=144,:)=[];
% for clump 8007005
trackinfo(~ismember(trackinfo.timeframe, 15:18),:)=[];Two tests will be made: a single iterative following and an iterative
cell following with the presence of clumps. The single following will be done
with the cells involved in clump 8002, studied in
shapeanalysis-log.md. The following inside the
clump will be done on clumps 8002 starting from frame 415, and 11010,
starting from frame 1.
This is allows the table to be better organised.
trackinfo(trackinfo.timeframe<418,:) = [];
aa = table2struct(trackinfo(1:length(clumplab):end,:));
jx=1:length(clumplab):size(trackinfo,1);
fields = {'X','Y','seglabel','track','finalLabel'};
for ix=1:length(jx)
for kx=1:length(fields)
auxvect = zeros(1,length(clumplab));
for qx=1:length(clumplab)
auxvect(qx) = trackinfo(jx(ix)+(qx-1),:).(fields{kx});
end
aa(ix).(fields{kx}) = auxvect;
end
aa(ix).clumpseglabel = aa(ix).seglabel(1)*(aa(ix).clumpcode>0);
end
trackinfo = struct2table(aa);
clear ix jx;The code developed in script_iterevolution.m,
is being written to address both the test case where a single, independent cell
is followed, as well as the actual practical case where a cell is followed
until it overlaps, and then the algorithm is run on it.
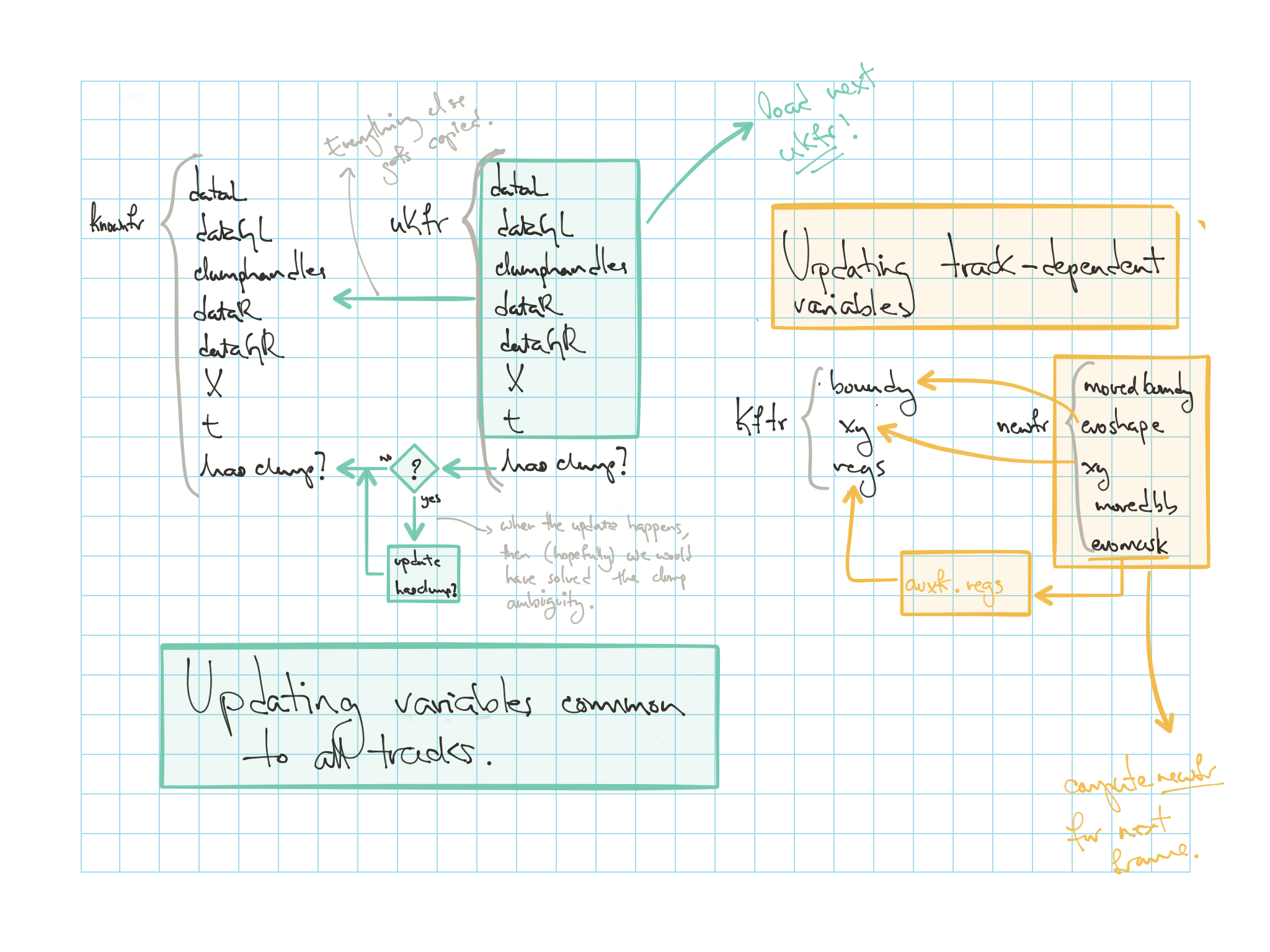
tk=1;
framet = trackinfo.timeframe(tk);
[knownfr] = getdatafromhandles(handles, filenames{framet});
knownfr.t=framet;
knownfr.hasclump = false;or (easier)...
[knownfr] = getCommonVariablesPerFrame(handles, trackinfo, wuc, ...
filenames{framet}, framet);kftr.regs = regionprops(zeros(size(knownfr.dataGR)), ...
'BoundingBox', 'Centroid', 'EquivDiameter', 'MajorAxisLength', ...
'MinorAxisLength');
kftr.boundy = cell(length(clumplab),1);
kftr.xy = zeros(length(clumplab),2);
for wtr=1:length(clumplab)
% K.F.Tf = Known Frames' TRacks
thisseglabel = trackinfo.seglabel(tk, wtr);
thiscell = knownfr.clumphandles.nonOverlappingClumps==thisseglabel;
regs = regionprops(thiscell, 'BoundingBox', 'Centroid', ...
'EquivDiameter', 'MajorAxisLength', 'MinorAxisLength');
boundy = bwboundaries(thiscell);
xin = trackinfo(trackinfo.timeframe==framet,:).X(wtr);
yin = trackinfo(trackinfo.timeframe==framet,:).Y(wtr);
kftr.regs(wtr) = regs;
kftr.boundy{wtr} = boundy{1};
kftr.xy(wtr,:) = [xin yin];
clear thisseglabel thiscell regs boundy xin yin
endtkp1 = tk+1;
frametplusT = trackinfo.timeframe(tkp1);
[ukfr] = getdatafromhandles(handles, filenames{frametplusT});
ukfr.t=frametplusT;
if trackinfo.clumpcode(tkp1) == wuc
ukfr.hasclump = true;
ukfr.clumpseglabel = trackinfo.clumpseglabel(tkp1);
ukfr.thisclump = (oneuk.dataGL==ukfr.clumpseglabel);
else
ukfr.hasclump = false;
endor much easier...
[ukfr] = getCommonVariablesPerFrame(handles, trackinfo, wuc, ...
filenames{frametplusT}, frametplusT);now, this one's easy....
[newfr] = nextframeevolution(ukfr, kftr, trackinfo, clumplab);plotBoundariesAndPoints(ukfr.X, newfr.movedboundy, newfr.evoshape, 'm-');
title(sprintf('Frame %d', frametplusT));It is in this part of the code where the variables will be disambiguated and saved back to the hard drive. Because this is
knownfr = ukfr;
if knownfr.hasclump == true
knwonfr.hasclump = false;
knownfr.clumpseglabel = [];
knownfr.thisclump = [];
endauxfr.regs = regionprops(zeros(size(knownfr.dataGR)), ...
'BoundingBox', 'Centroid', 'EquivDiameter', 'MajorAxisLength', ...
'MinorAxisLength');
auxfr.boundy = newfr.evoshape;
auxfr.xy = newfr.xy;
for wtr=1:length(clumplab)
thiscell = newfr.evomask(:,:,wtr)>0;
regs = regionprops(thiscell, 'BoundingBox', 'Centroid', ...
'EquivDiameter', 'MajorAxisLength', 'MinorAxisLength');
auxfr.regs(wtr) = regs;
end
kftr = auxfr;
clear auxfr;As mentioned before, clump wuc=8002 was analysed from frame 418 till frame
468 because it is a range where the cells involved do not overlap, an can be
followed on their own, testing the method's basic premise.
The parameters of the activecontour function were defined, for each
frame, as:
| method | iter | smoothf | contractionbias |
|---|---|---|---|
| Chan-Vese | 50.00 | 2.00 | -0.10 |
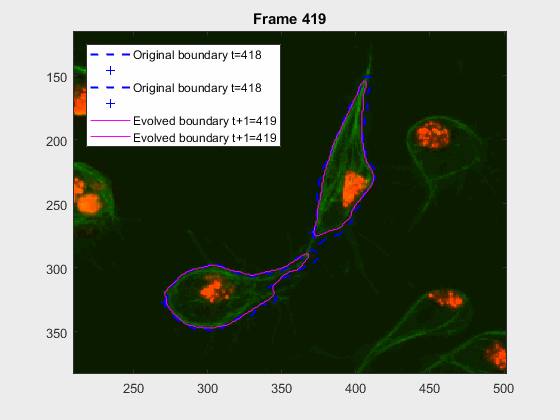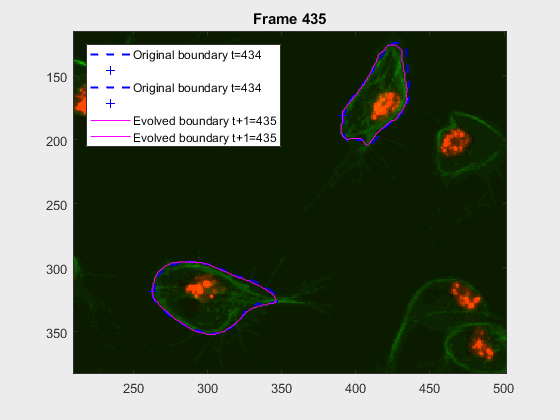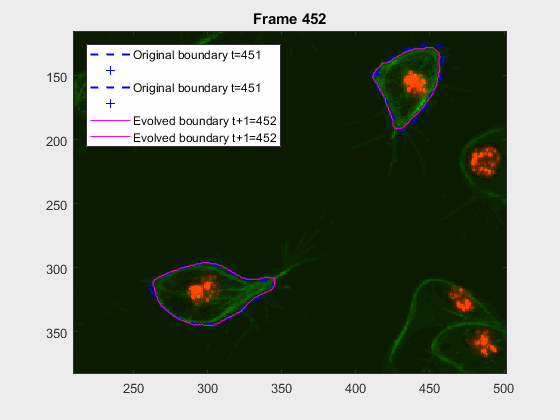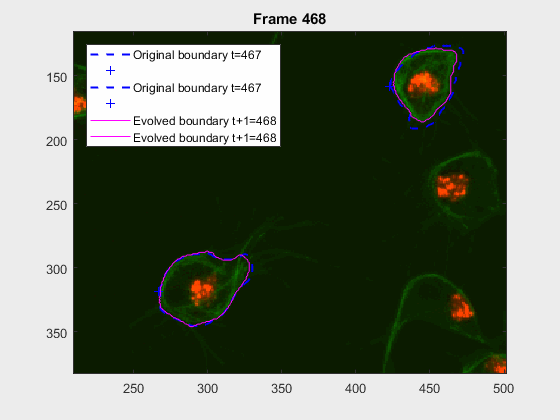
It can be seen how the cells are followed successfully, which does not impress anyone, because of the initial positions of the cells.
This experiment takes place in the frames leading up to 417, where the
clump wuc=8002 is formed. It is a good initial example to try and lead up the
following of a pair of cells that eventually clash into a clump, but then
separate immediately on the next frame.
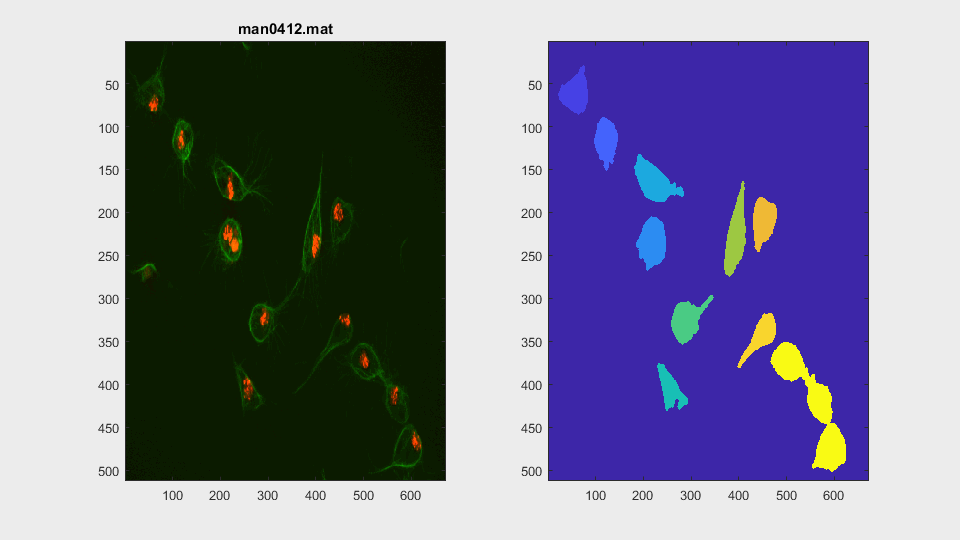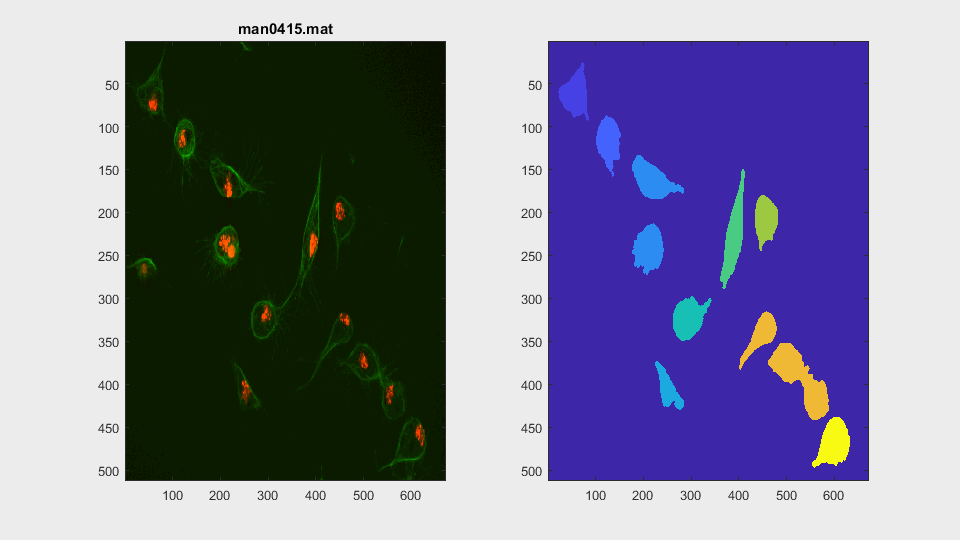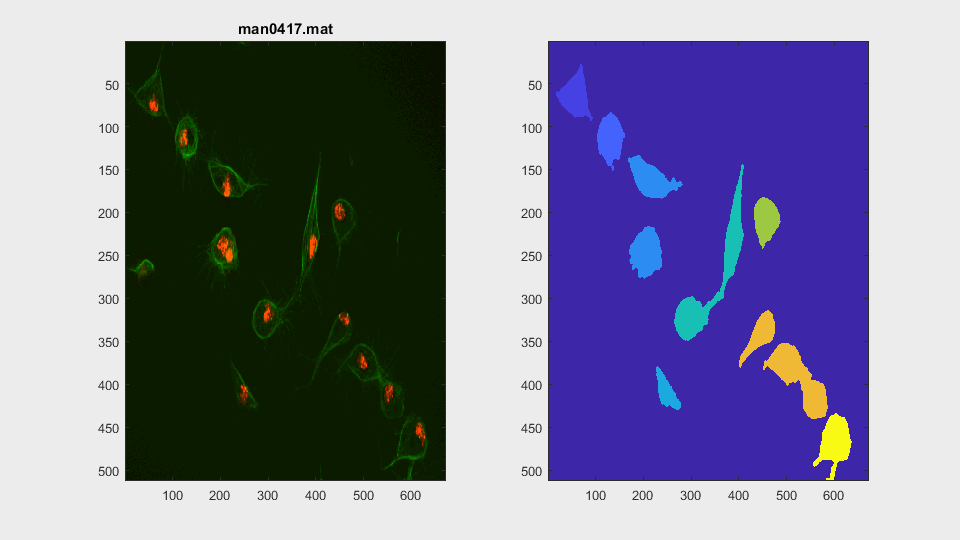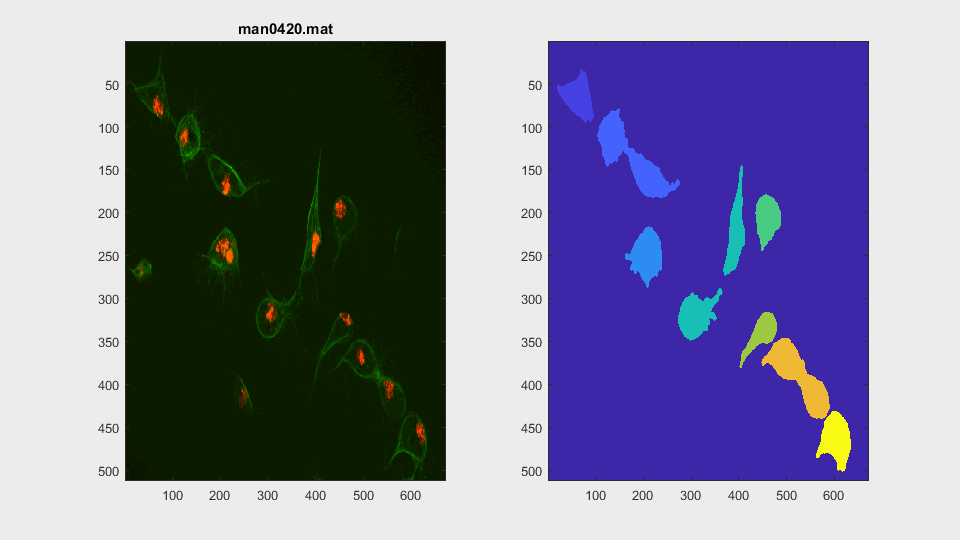
The parameters of the activecontour function were defined, for each
frame, as:
| method | iter | smoothf | contractionbias |
|---|---|---|---|
| Chan-Vese | 50.00 | 2.00 | -0.10 |
And the results:

This is a very easy clump, as it does not involve overlapping, however it shows the ability of the method to switch from analysing a frame with or without clump.
This is one of the hardest clumps, because the shapes overlap completely and the previous information is not enough to do anything.
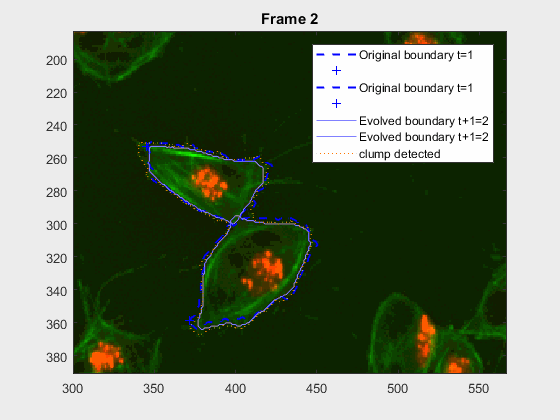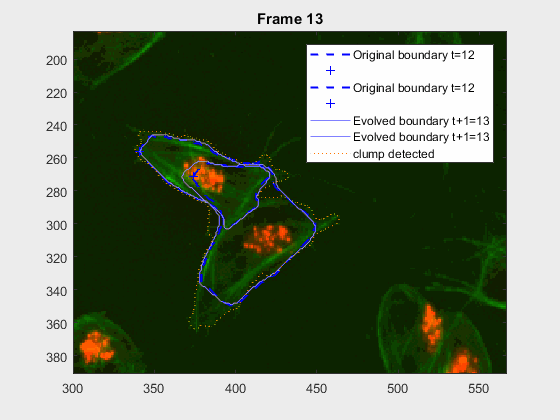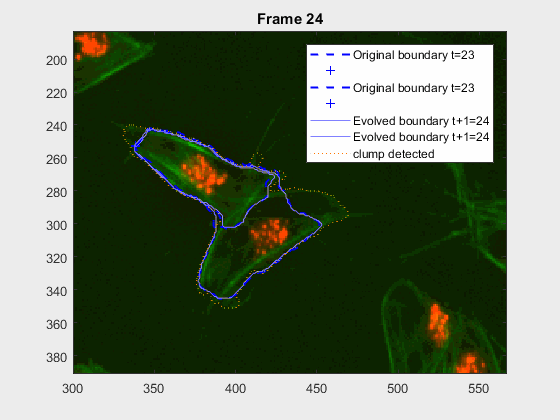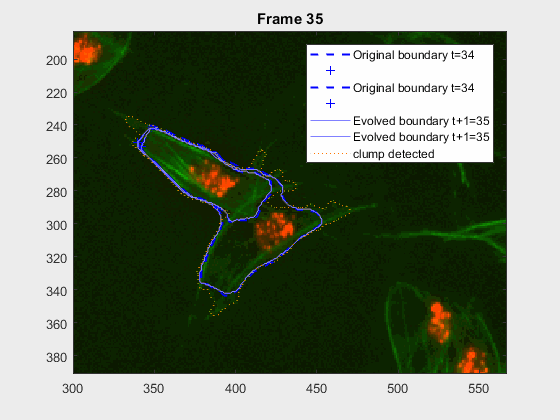
This shows that the method should probably have some way of detecting these types of transitions. We could very easily detect when the wrong cell becomes a clump again. While I can imagine some post processing helping, I do believe this approach lacks the information of the shape to stop the contour to take up difficult shapes.
First, let's look at an example:
Not only disambiguate, but also setting the path for updating the .mat files
that contain the segmentations. Solving this clump will lead into solving
clump 8007.

The following step from this is to update the information on dataGL
to rid the clump.
We define an easy clump as one which does not contain actual overlapping
of the cells, but that the detection through thresholding, or the morphology
steps when segmenting the green channel caused a clump where there was none.
Separating this type of clump could be easily done by updating the variables
present in ukfr.dataGL and then generate the necessary variables to be saved
in the corresponding filename{someframe} that exists in handles.dataLa.
Thus, the next time this frame is loaded, it will contain the appropriate
information about the clump.
The information on the clumps should also be addressed.
Such an update should occur in part
4.1 of
the algorithm. In here, the variable .hasclump is tested and with a
positive result, the variables are then changed.
Straightforward changes: dataGL, clumphandles
An assumption is made for the cells not overlapping yet (after all, this is the case of an easy clump).