-
Notifications
You must be signed in to change notification settings - Fork 0
scr iterevol areacontrol
This log file is a continuation of the file
scr-iteravolution-vs-leaking.md.
The focus of this (hopefully small) log file is to address some issues
that occur with the segmentation in the previous file regarding the
shape of the cell being segmented and the control we can realistically
have over the MATLAB function activecontour.m.
We know about the parameters of activecontour.m that can be addressed,
they involve:
-
Method: Choose from
'Chan-Vese'and'edge' -
Iterations: How many times the program is run. Typical values used here
are
options.iter=25andoptions.iter=50. -
Smooth Factor: Parameter that forces the contour to be soft, smooth
or regular. Typical values used in this application are
options.smoothf= 1.5andoptions.smoothf= 2, when the method used isChan-Vese. - Contraction Bias: Is the tendency of the contour to expand or shrink.
This function does not allow a finer control over the parameters, the evolution or the finer details (restricting size, volume or area of the contours).
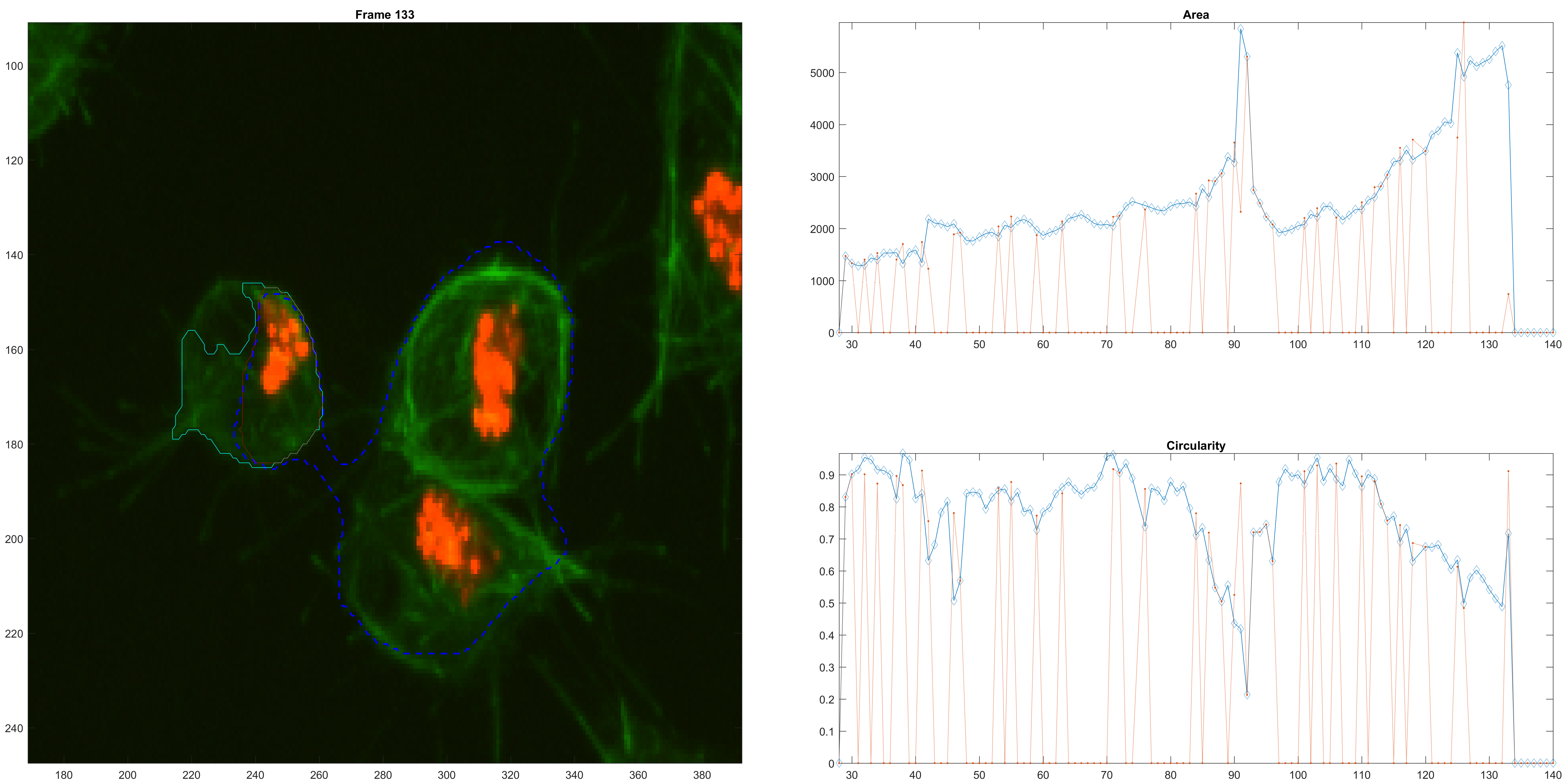
Upon trying to compute the method for all the independent cells, some mistakes
in the evolution of the shape were found. And because of the nature of the
evolution, where a a frame at t(k+1) is created from the previous
t(k+1)=tk+1, the errors in segmentation in one frame can be passed
on to the subsequent ones.
For this experiment, the cell at tracklabel=2 was examined in two different
experiments: from frames 28:300 and 202:300. In this case, there were many
objectives:
- Follow a cell over long span of frames, and see how the following mechanism would cope.
- Follow a single cell even though it might go through clumps.
- See how different would the cells be segmented when starting at different
points.
- How would cell
2look at frames>230when starting at frame28? - how would it look upon starting at frame
202?
- How would cell
Some results were found, the .gif files appear below

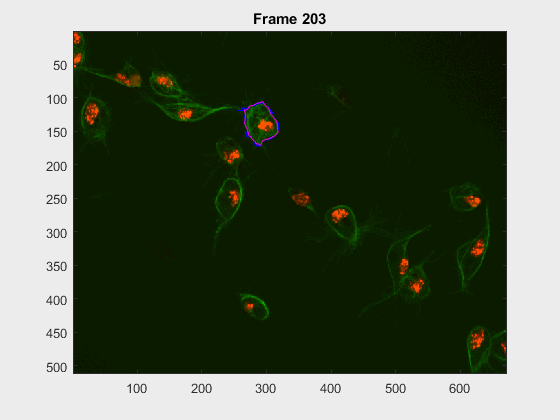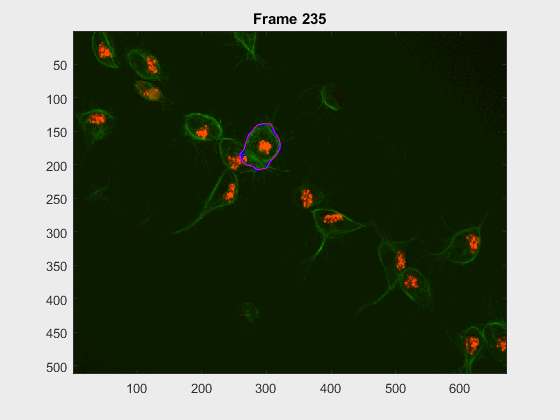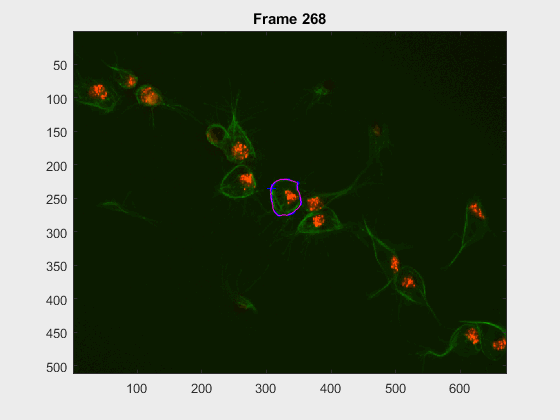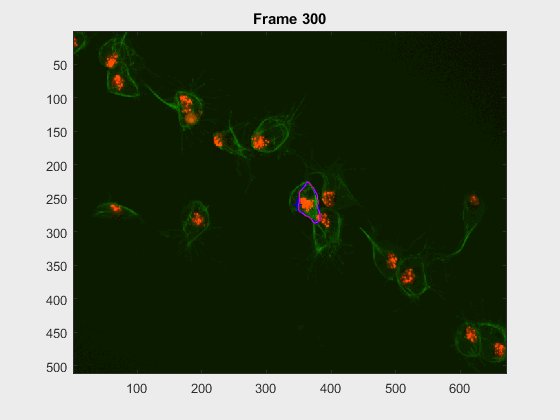
The method is able to cope with the following of the cell, but
- Segmentation errors are passed on between frames and only in specific scenarios does the segmentation fixes itself.
- In frames where a clump is found, the segmentation falls into even more traps or leaks.
- Noticeably, the segmentation from frames
230:300becomes pretty much the same, even with the very different segmentation results at frame202. The measurements were taken, just as in the script filescr-iteravolution-vs-leakingand the measurements matched after some frames (not shown).
For this section of the log, an initial analysis is made with the
activecontour function. A generic example is tried.
I = imread('toyobjects.png');
figure(1)
imagesc(I);
mask = roipoly;
figure(2)
imagesc(mask);
title('Initial MASK');Now the parameters, and the actual function:
acopt.method = 'Chan-Vese';
acopt.iter = 200;
acopt.smoothf = 1.5;
acopt.contractionbias = 0;
acopt.erodenum = 5;
evomask = activecontour(I, mask, acopt.iter, acopt.method, ...
'ContractionBias',acopt.contractionbias,'SmoothFactor', acopt.smoothf);
figure(3)
plotBoundariesAndPoints(I, bwboundaries(mask), bwboundaries(evomask), 'm-');- Run
initscriptso we have access to thehandlesstructure. - Images where some problems can be found for cell
2are frames43, 96.
The painless way to run tests:
initscript;Then just load everything:
a = getdatafromhandles(handles, filenames{43})
clumplab = 2;
wuc = 2;
trackinfo = [tablenet(ismember(tablenet.track, clumplab),[5 1 2 11 13 14]) ...
clumptracktable(ismember(tablenet.track, clumplab),:)];Plot the image (with current segmentation) and get a different one with
roipoly, that will simulate a previous step:
plotBoundariesAndPoints(a.X,...
bwboundaries(a.dataGL==trackinfo(trackinfo.timeframe==43,:).seglabel));
mask = roipoly;Get the parameters for the activecontour,
acopt.method = 'Chan-Vese';
acopt.iter = 200;
acopt.smoothf = 1;
acopt.contractionbias = -0.25;
acopt.erodenum = 5;finally, just run the routine:
evomask = activecontour(a.dataGR, mask, acopt.iter, acopt.method, ...
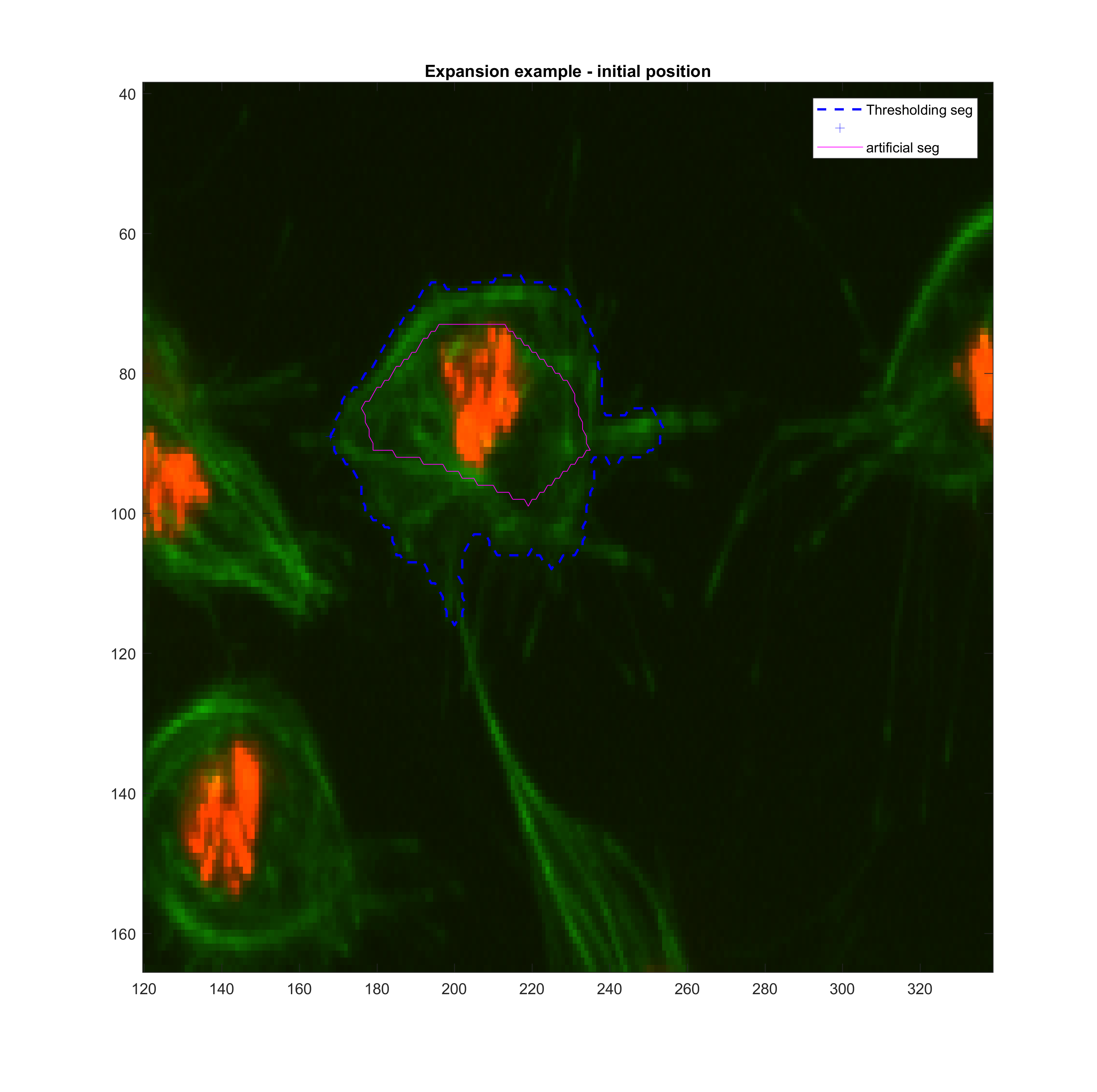
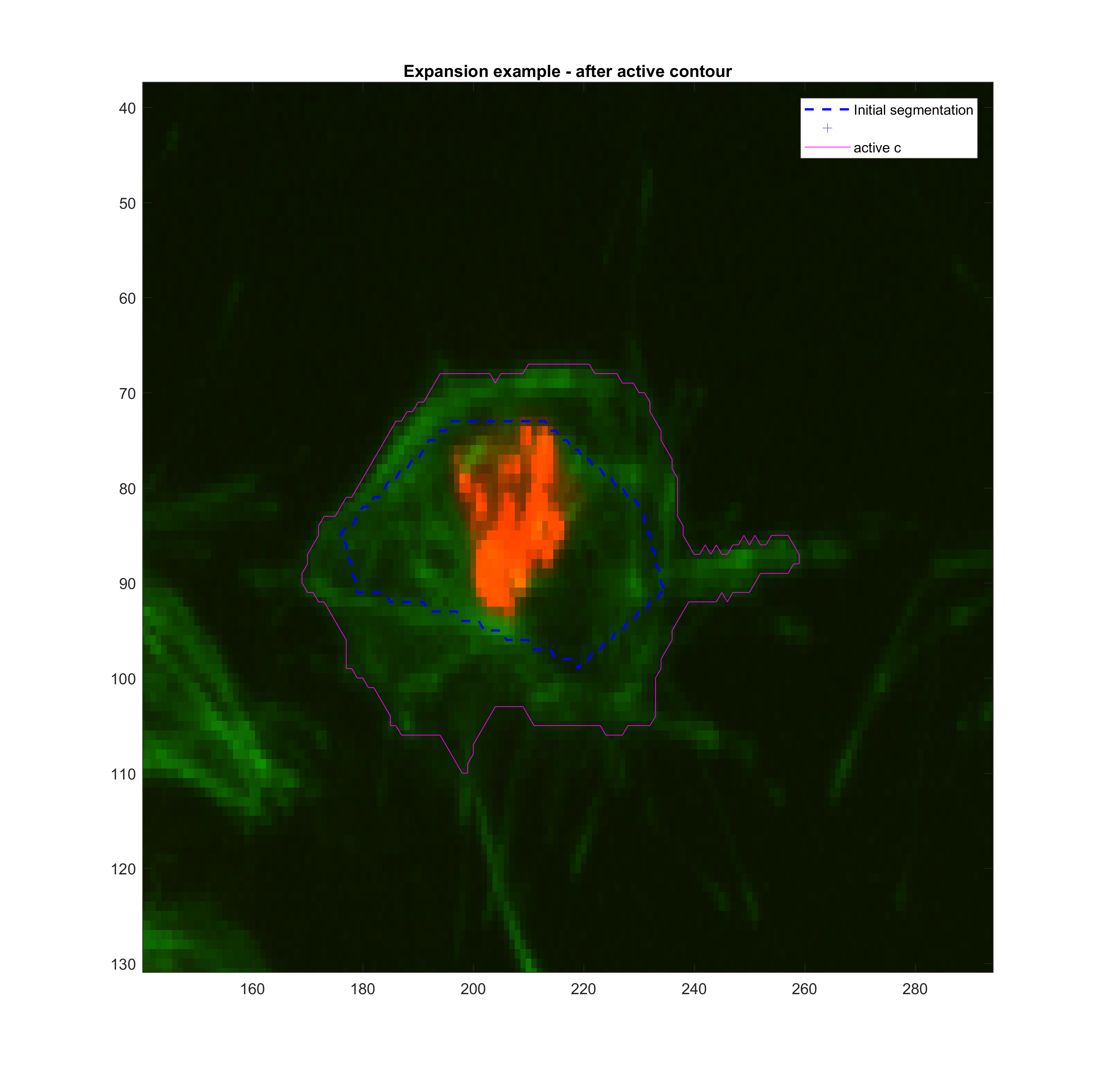
'ContractionBias',acopt.contractionbias,'SmoothFactor', acopt.smoothf);The same funciton roipoly was used to simulate a smaller or larger segmented
area for frame t(k-1). Then, various tests with the function activecontour
were done in order to get a sense
Growing
| method | iter | smoothf | contractionbias | erodenum |
|---|---|---|---|---|
| Chan-Vese | 200.00 | 1.00 | -0.25 | 5.00 |
|
Shrinking
| method | iter | smoothf | contractionbias | erodenum |
|---|---|---|---|---|
| Chan-Vese | 100.00 | 1.25 | 0.10 | 5.00 |
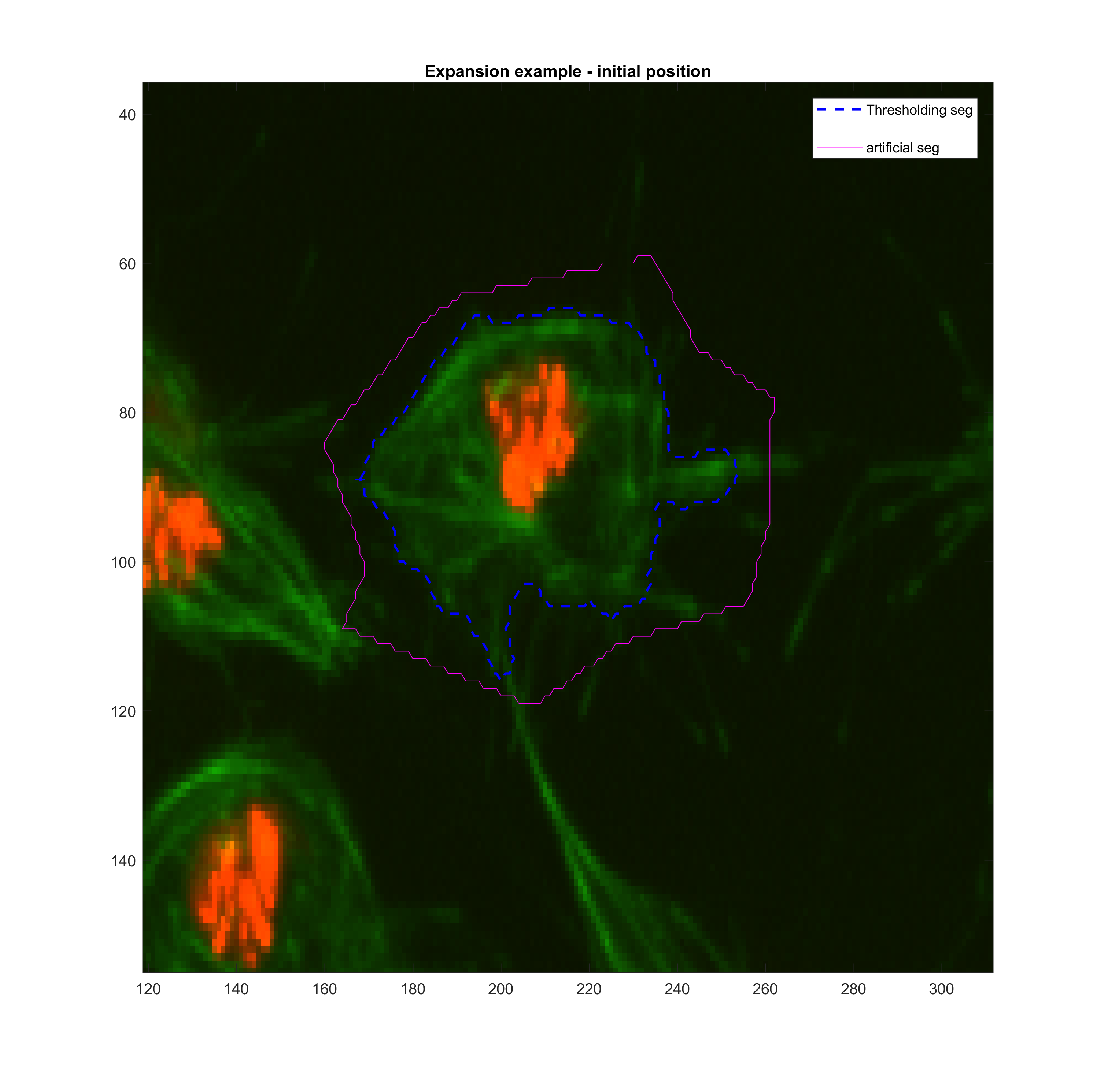 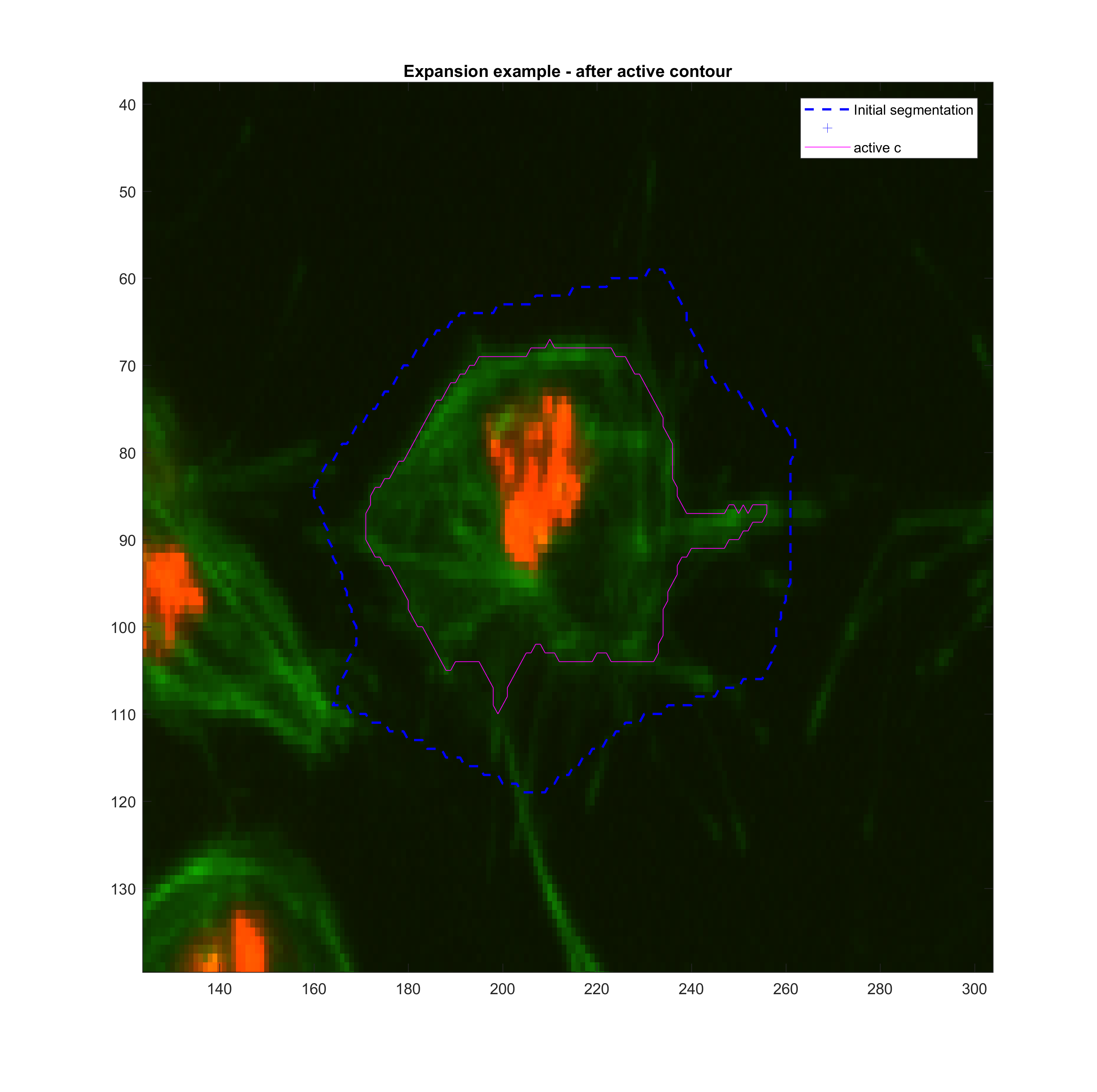
|
These parameters may need to be changed when addressing clumps.
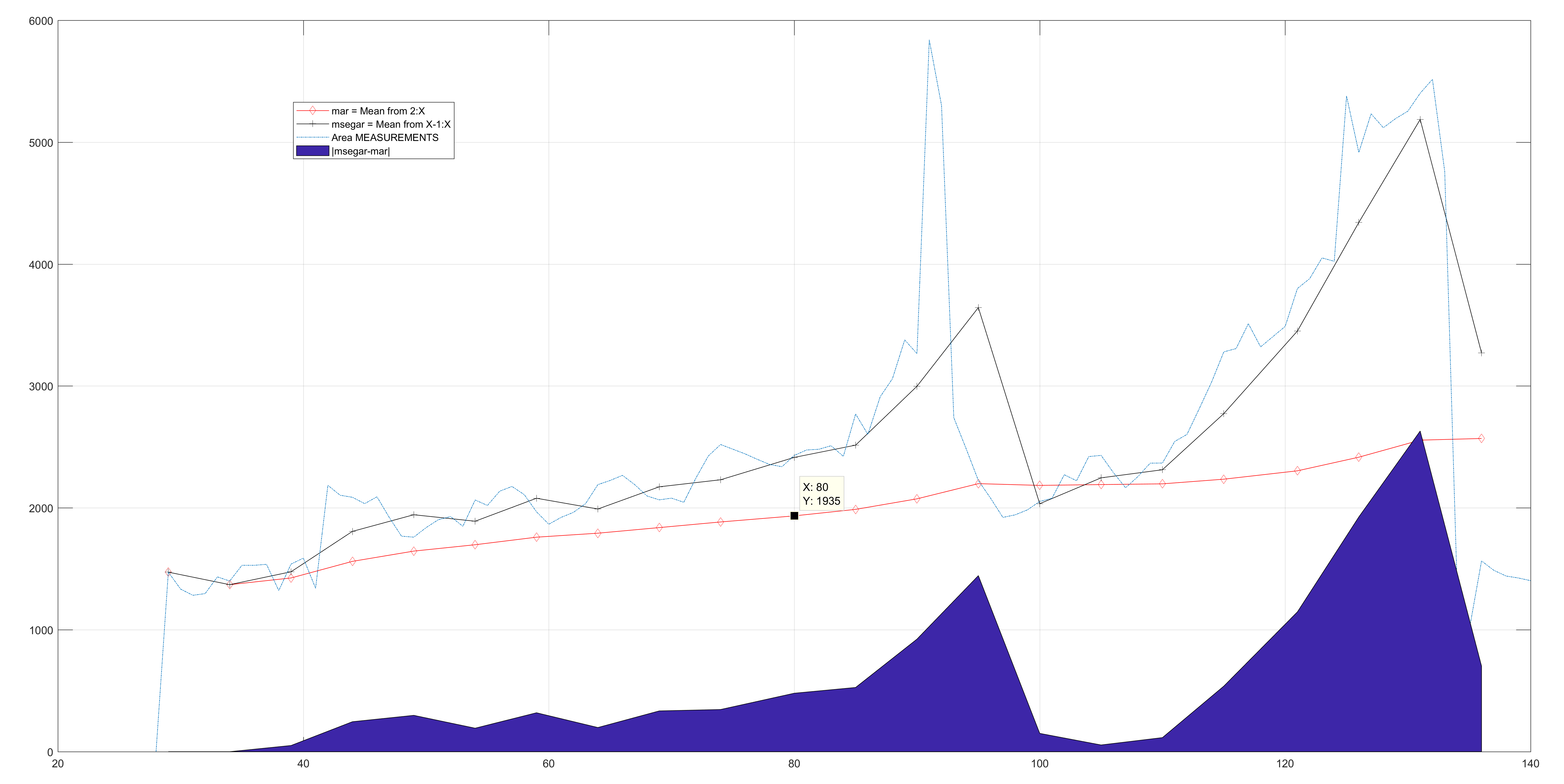
The approach described before was implemented in a crude way. At each step, the size (Area) of the new frame is compared with the previous size. A large change results in the adjustment of the
An example of size correction.
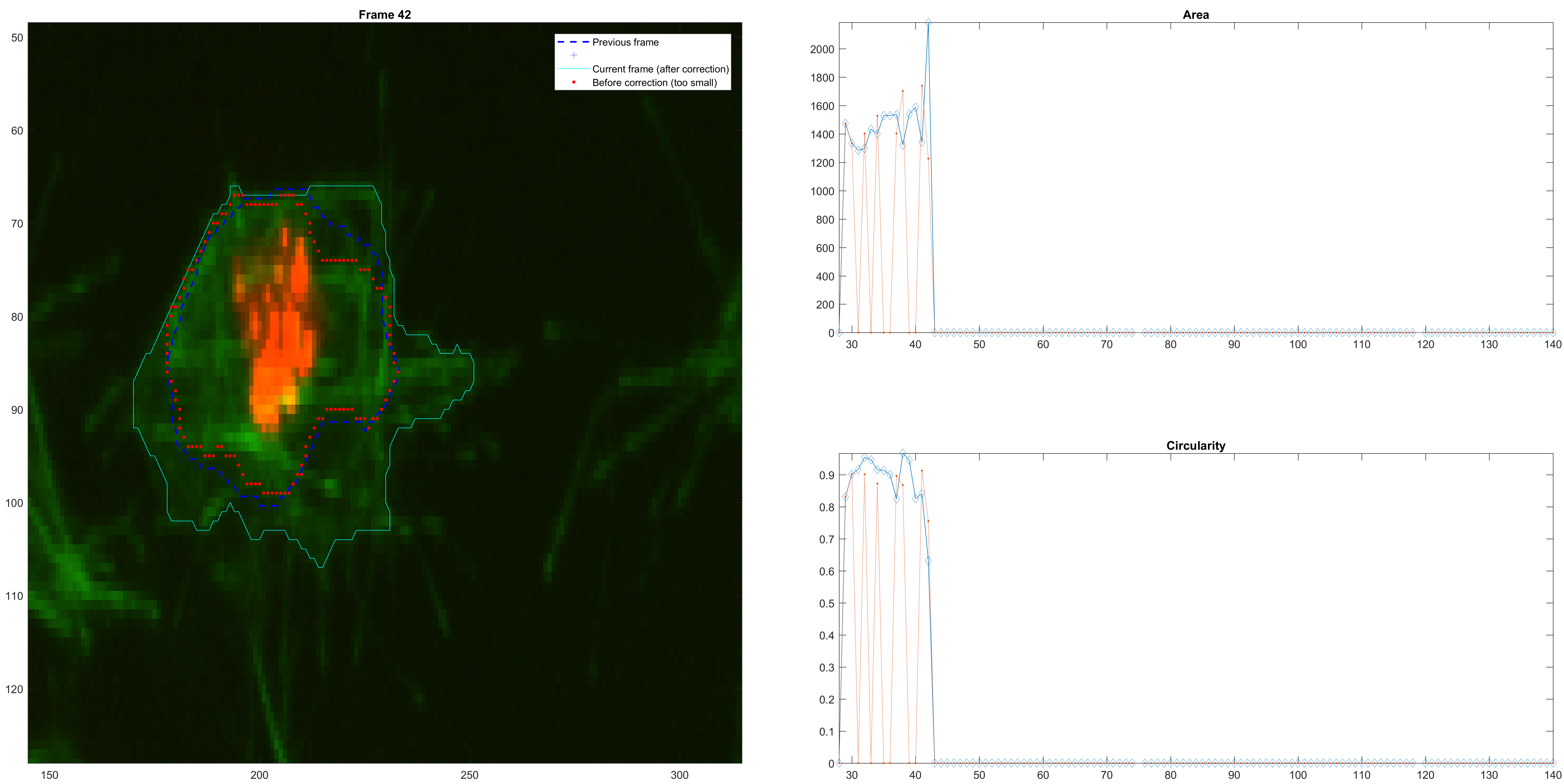
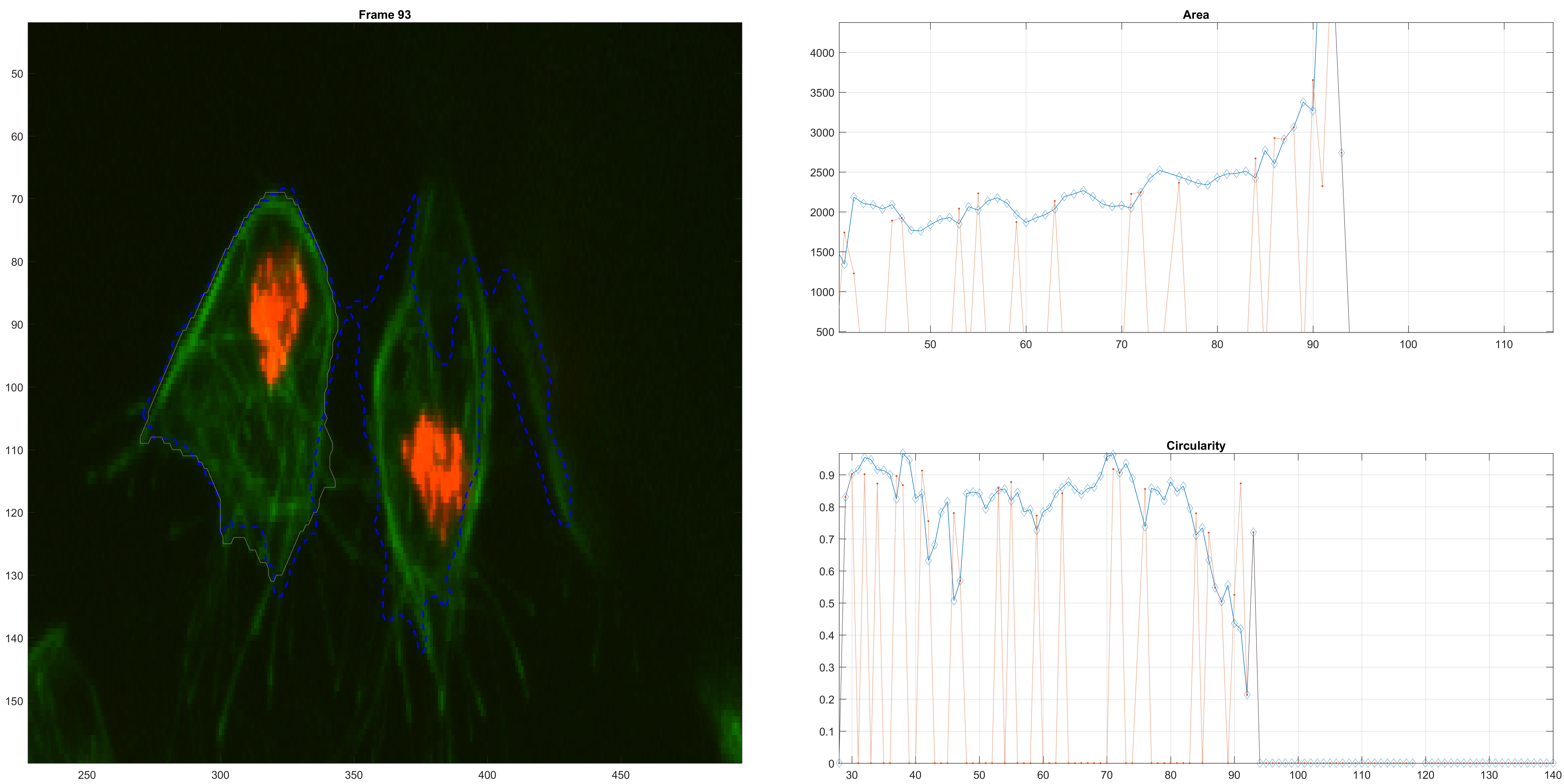
Right before a clump.
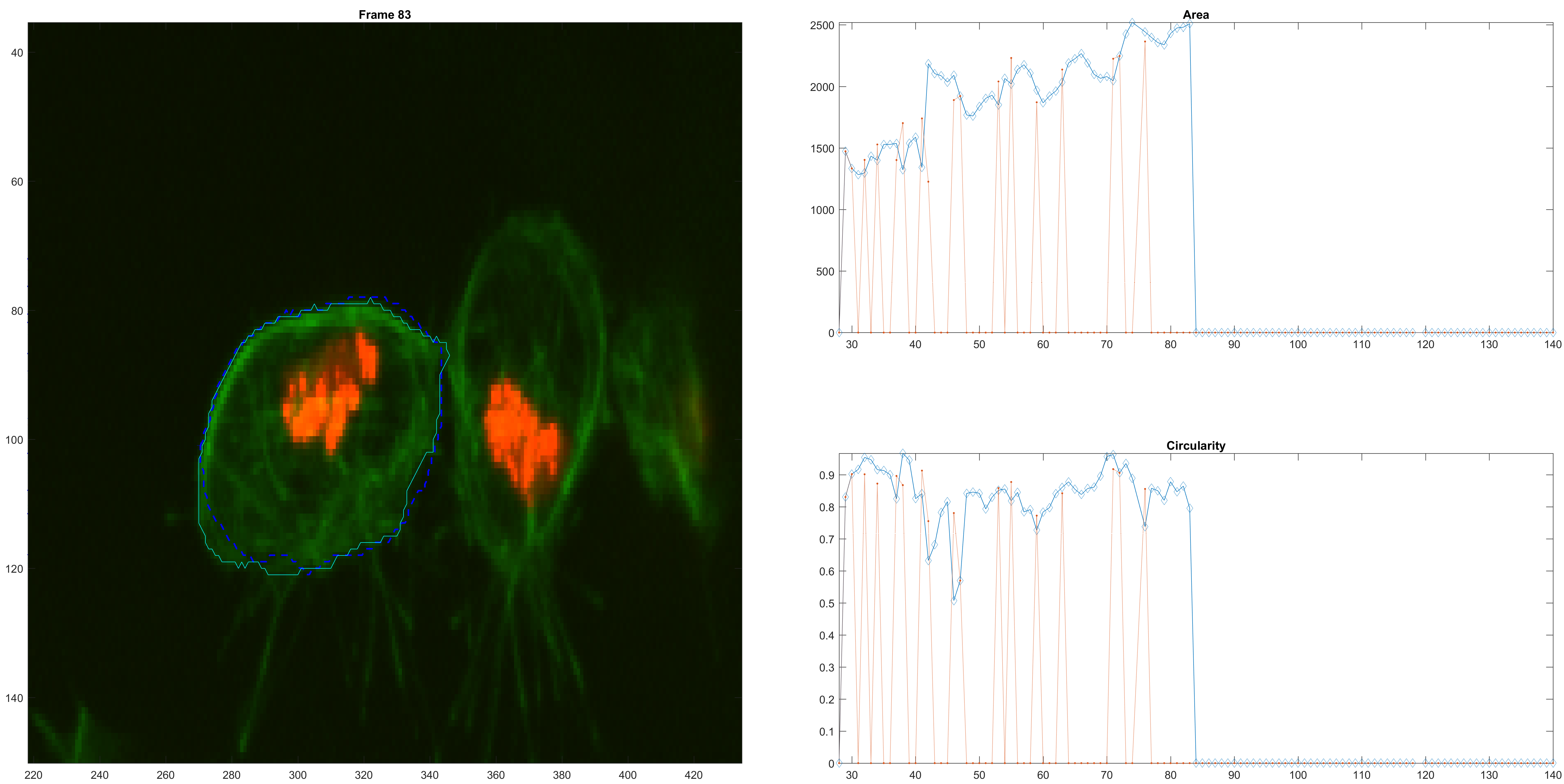
A clump confusion.
A bigger confusion.
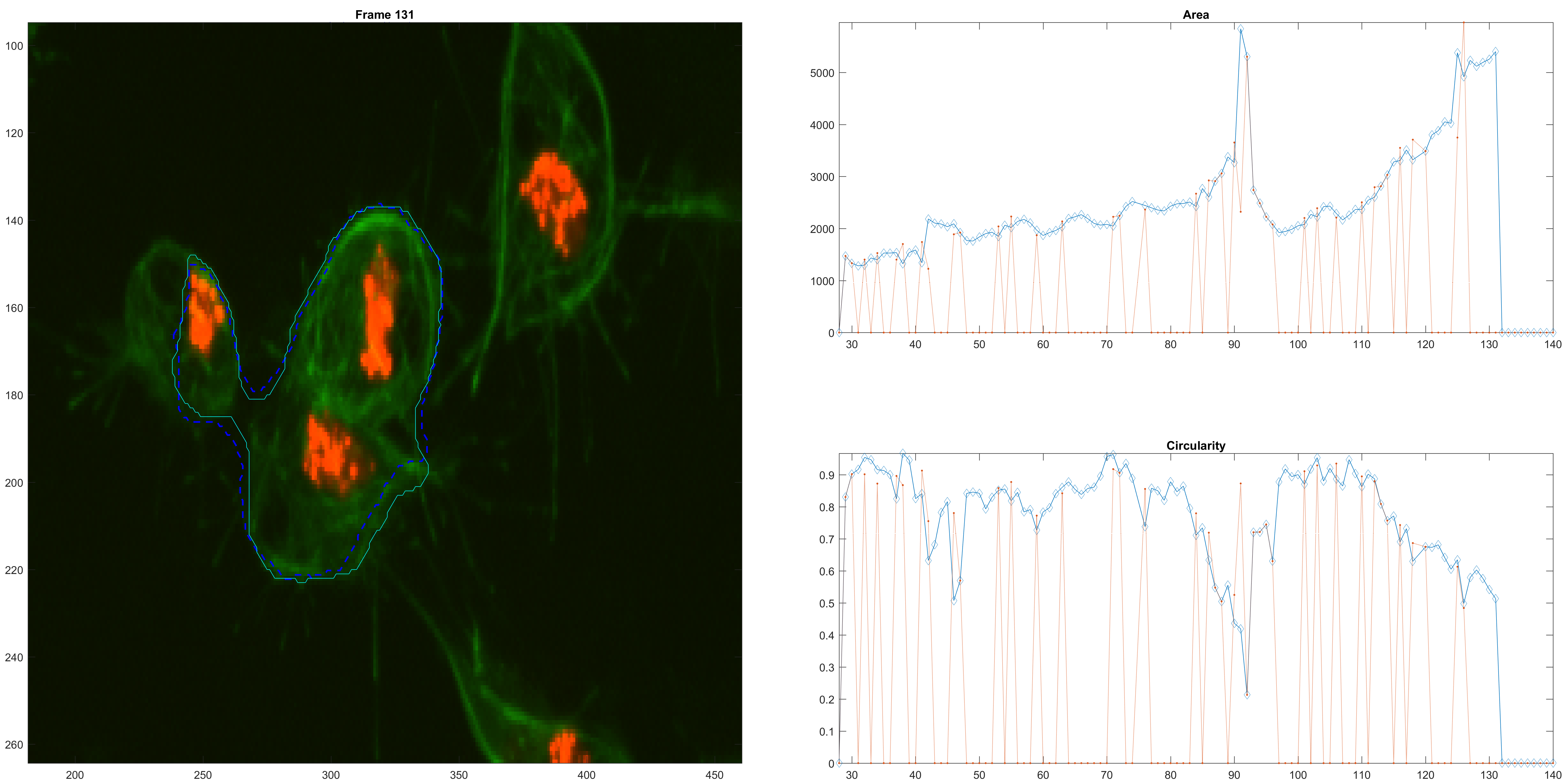
A loss of the original cell because of the confusion.