-
Notifications
You must be signed in to change notification settings - Fork 0
scr iteravolution vs leaking
This log file follows the development described in the
shapeevolution-iterative.md file.
The script file
script_iterevolveVSleaking.m
is described in this case, which is a condensed version of
script_iterevolve.m
that allows to make the detection and handling of leaking segmentation
more straightforward.
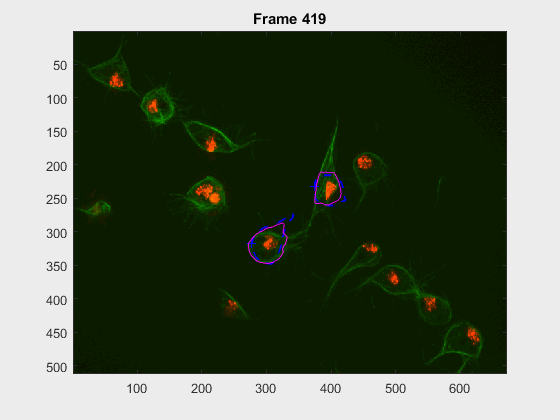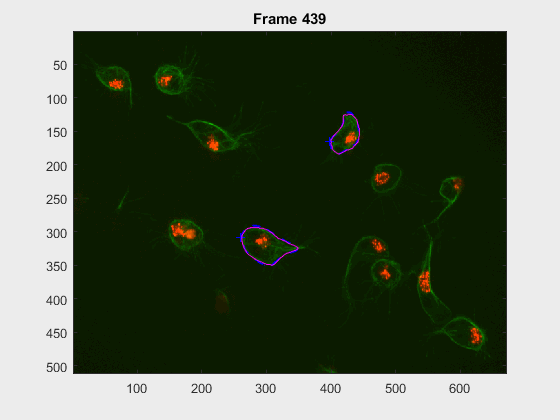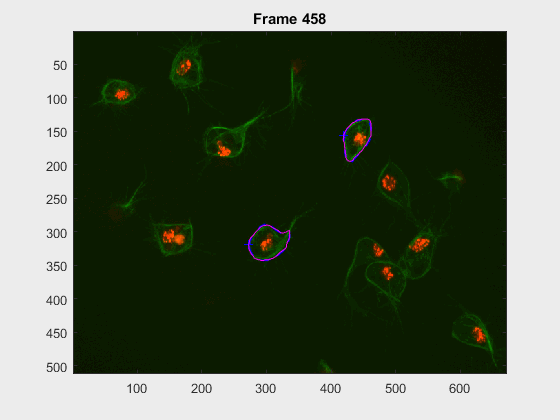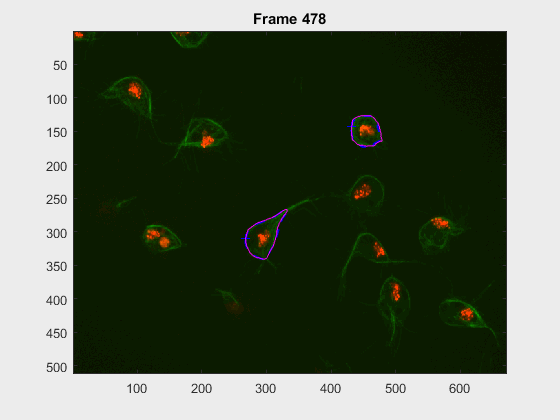
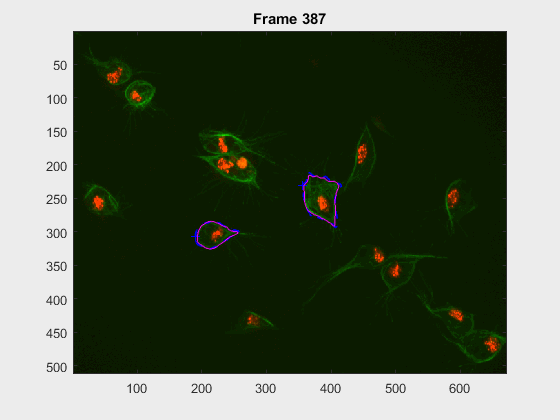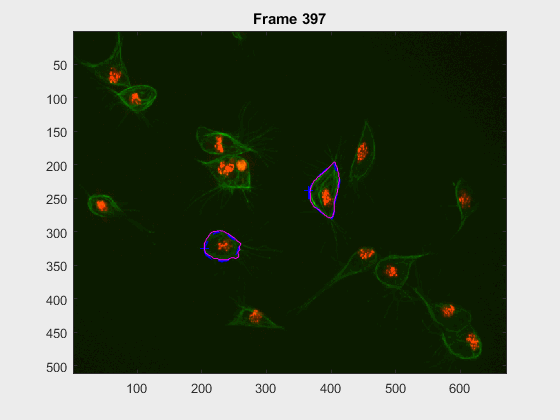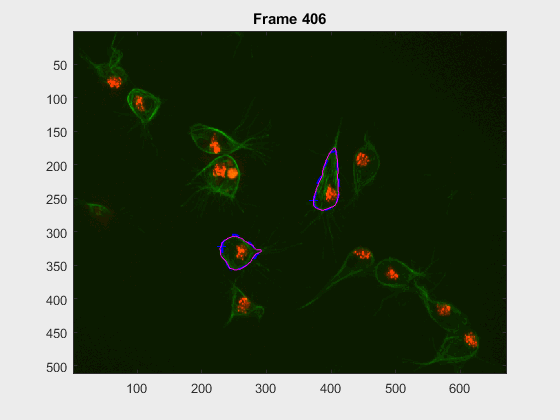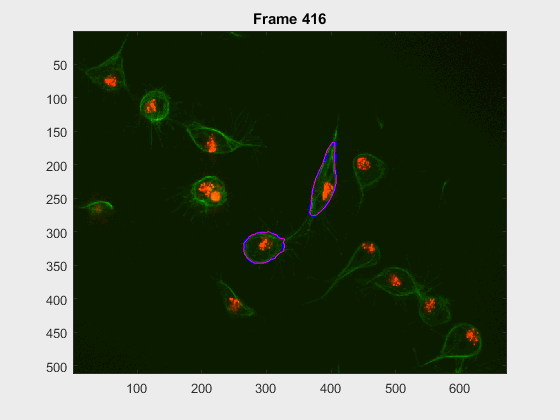
The following image makes the problem explicit:
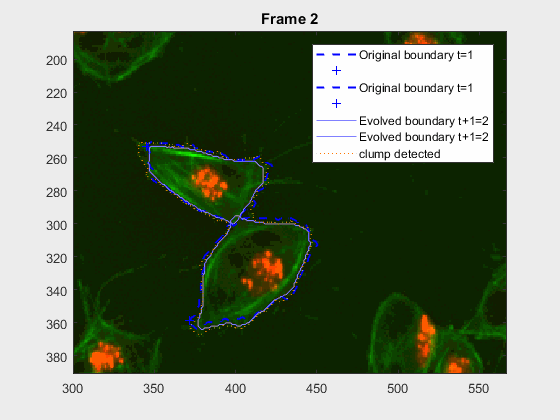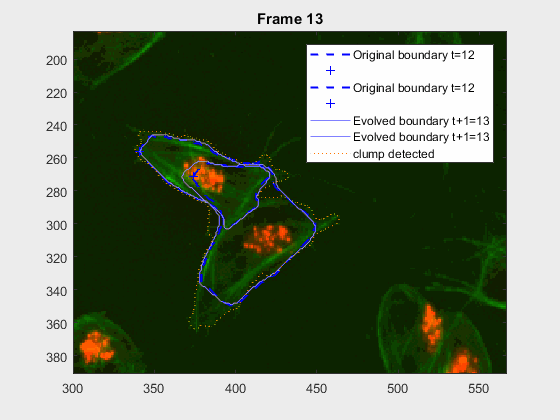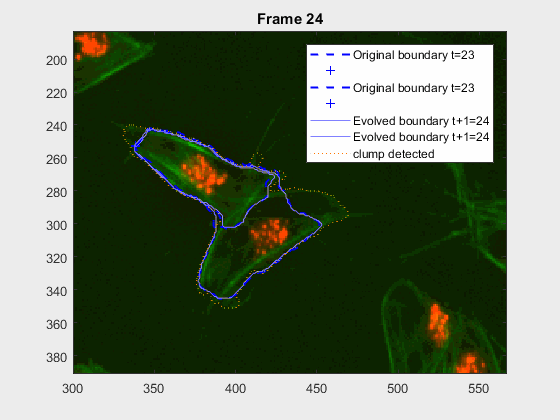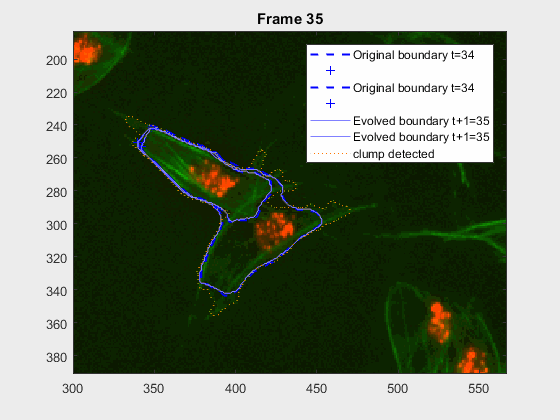
The code so far can handle easy clumps, but in some cases, the segmentation
leaks and takes another cell. In this log (and .m) file(s) the priority
falls into detecting
The code developed in script_iterevolve.m
is displayed, in a condensed form. All of the functions used are part of the
@macrosight package.
% 1. Load known frame
tk=1;
framet = trackinfo.timeframe(tk);
[knownfr] = getCommonVariablesPerFrame(handles, trackinfo, wuc, ...
filenames{framet}, framet);
% 2. Compute the tracks' variables in the known frame
[kftr] = getKnownTracksVariables(knownfr, trackinfo, clumplab, tk);
%% 3.1 Load the unknown frame
tkp1 = tk+1;
frametplusT = trackinfo.timeframe(tkp1);
[ukfr] = getCommonVariablesPerFrame(handles, trackinfo, wuc, ...
filenames{frametplusT}, frametplusT);
% 3.2 Evolve
acopt.method = 'Chan-Vese';
acopt.iter = 50;
acopt.smoothf = 1.5;
acopt.contractionbias = -0.1;
acopt.erodenum = 5;
[newfr] = nextframeevolution(ukfr, kftr, trackinfo, clumplab, acopt);
% 3.4 Update
% 3.4.1 Update knownfr
[knownfr, kftr] = updateKnownFrame(ukfr, newfr, clumplab);
if ukfr.hasclump == true && false % change to true for updating info to disk
update2disk(handles, knownfr, newfr, wuc);
end
framet = frametplusT;
tk = tk+1;Two shape tests will be conducted to try and identify the calculations
necessary to detect a potential leaking with the segmentation, and maybe
provide a better solution to the problem. At the moment, the function
activecontour is being used naively, if the segmentation provided by it
can be assessed and fixed in the case of leaking, then A better approach, a
dynamic tweak in the parameters or disregarding one sector of the segmentation
altogether might be implemented.
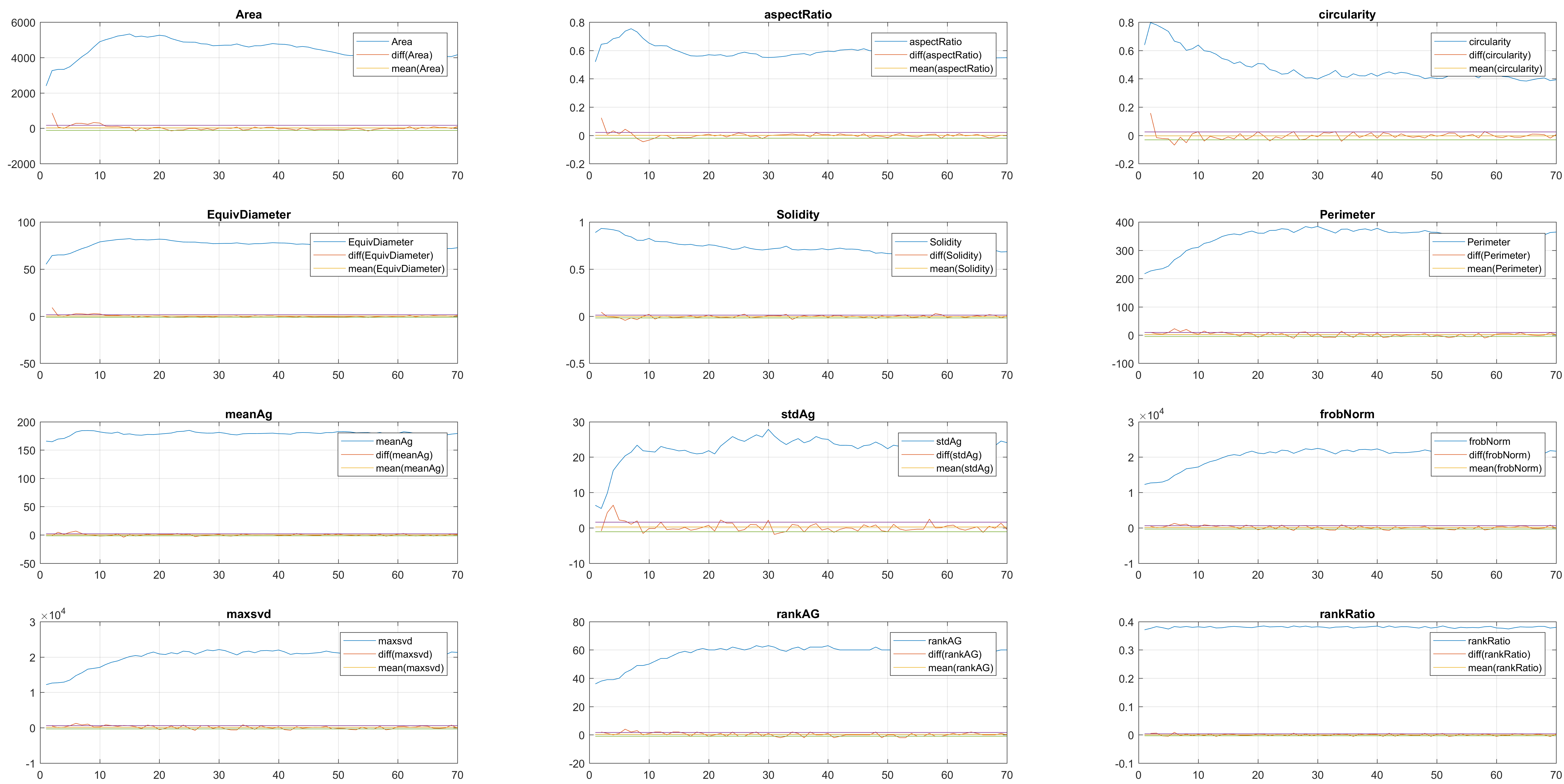
There are two major proposals in this area. The first is to calculate some traditional shape parameters like of each of the cells, at each segmentation point. Next, a description of each parameter is presented and how it is calculated:
-
Areacovered by the segmented pixels after the shape evolutionnewfr.evolmask. This is computed by Matlab'sregionprops. aspectRatio = MinorAxisLength./MajorAxisLength;circularity = (4*pi).*(Area./(Perimeter.^2));-
EquivDiameterThis is computed by Matlab'sregionprops. -
SolidityThis is computed by Matlab'sregionprops. -
PerimeterThis is computed by Matlab'sregionprops.
The other approach to find leaking in a segmentation is to compute the anglegram of each cell involved in the overlapping. My hypothesis is that this way, not only will we come to the detection of a leak, but also the right positions where the segmentation can be ignored, because it does not follow a normal curvature.
meanAg = mean(candyhandles.angleSummaryVector);stdAg = std(candyhandles.angleSummaryVector);frobNorm = norm(anglegram, 'fro');maxsvd = max(svd(anglegram));rankAG = rank(anglegram);rankRatio = rankAG/size(anglegram,2)
Both tests will start by testing a couple of cases where there is no overlap
and the cells involved are just moving in time. Then, the tests will move on
to clumps wuc=8007 and wuc=11010, which do overlap.
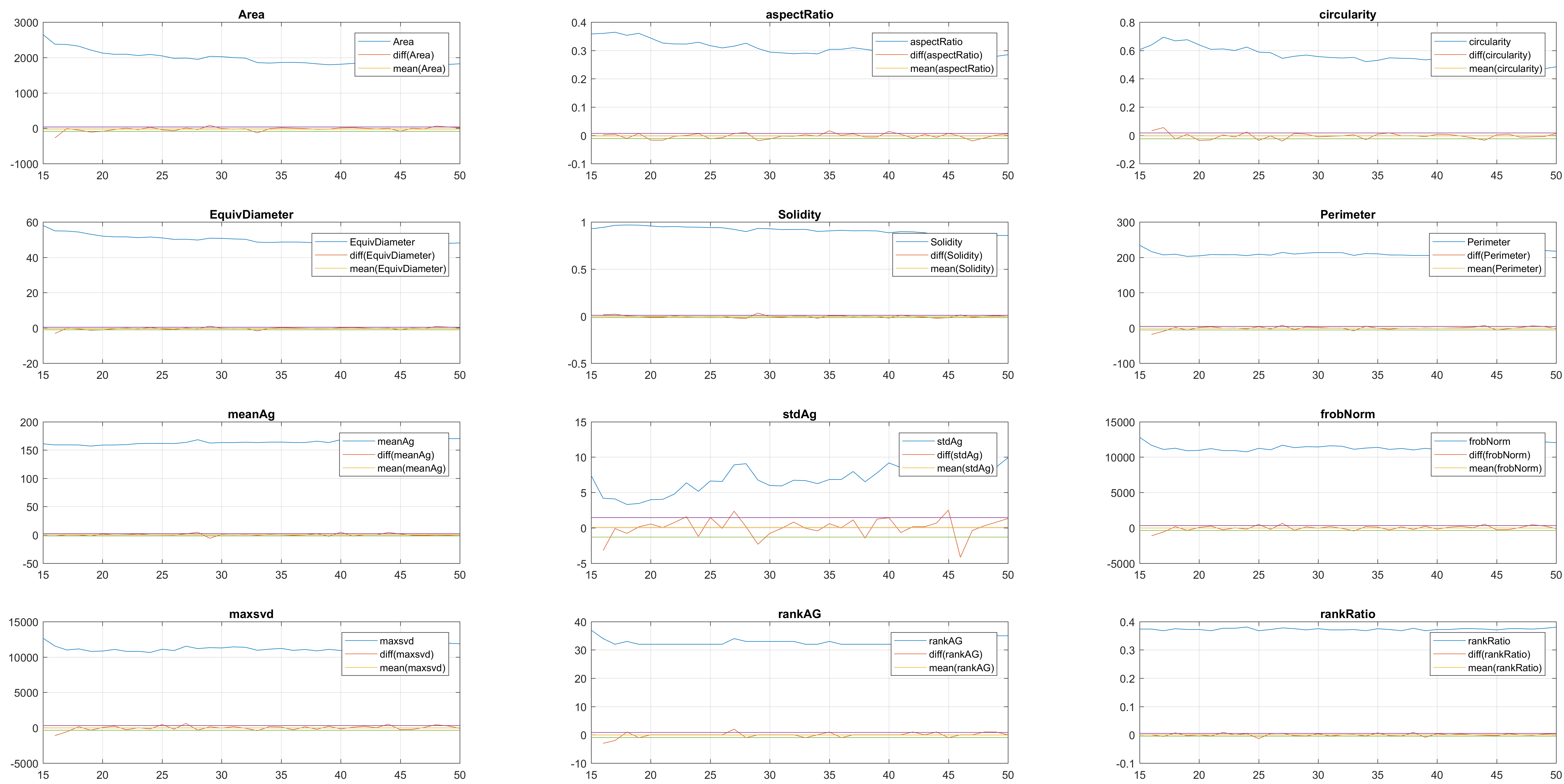
In this case, two non-overlapping cells were followed from wuc=8002 to
assess the different measurements described before. The cells assessed in this
case are as follows:
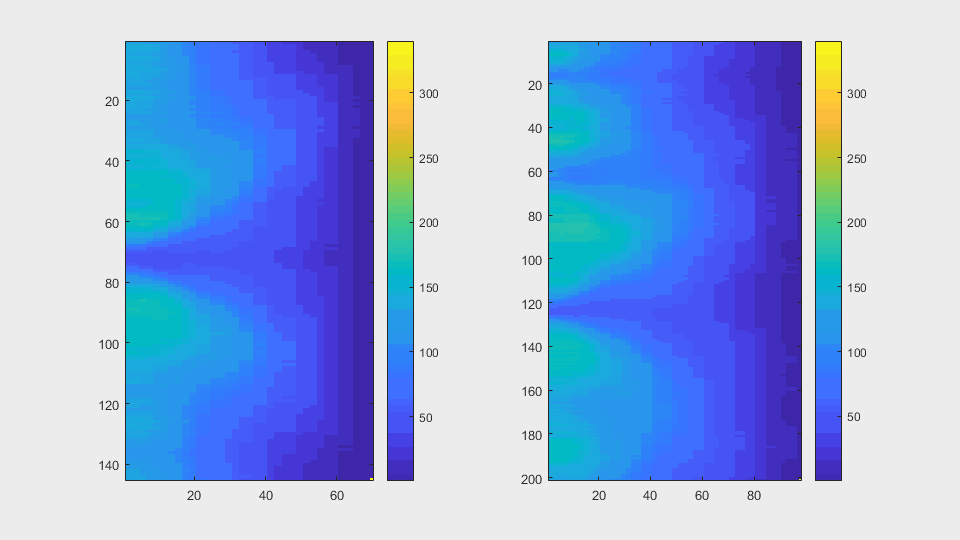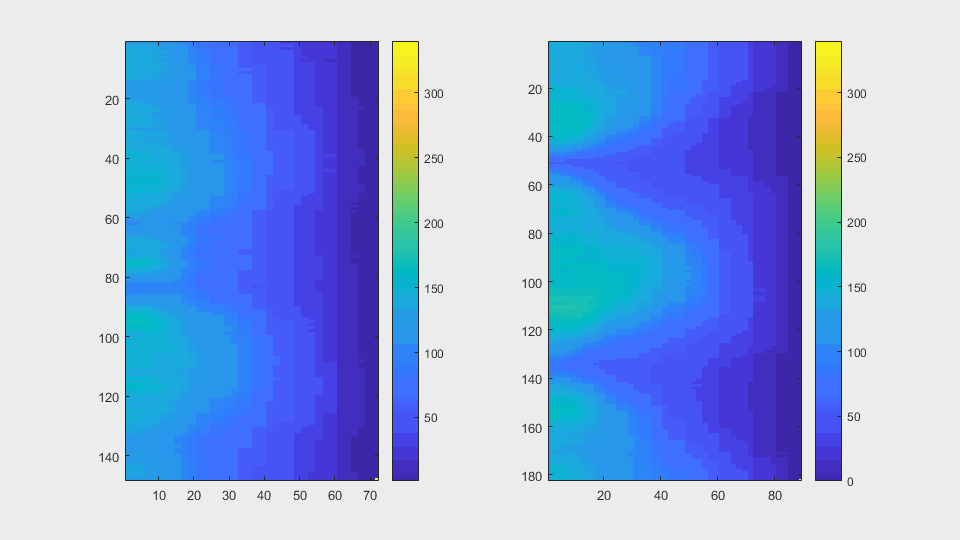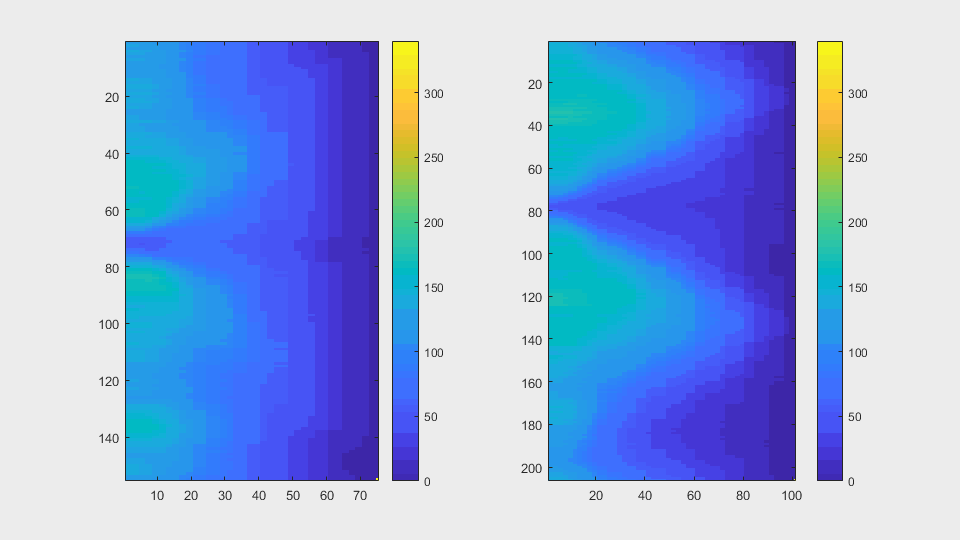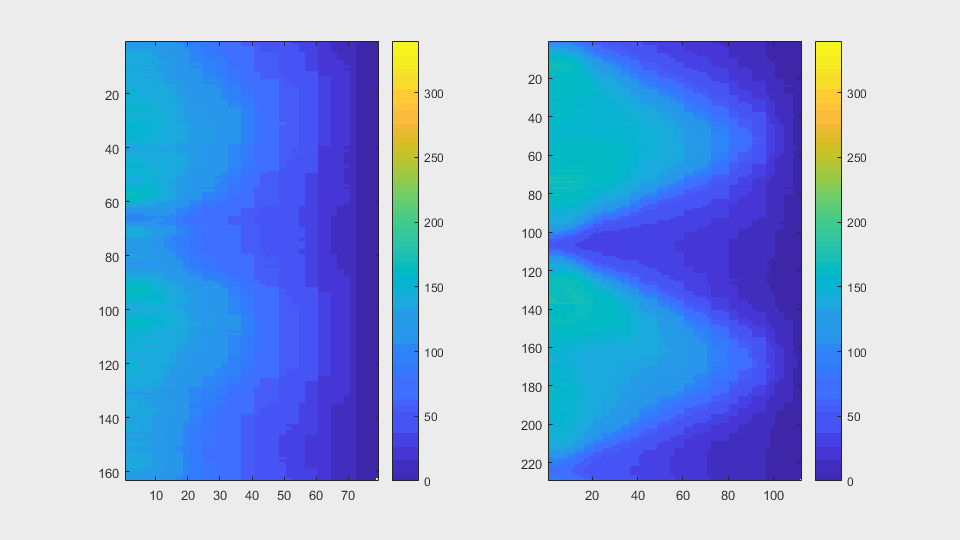
The anglegrams of both cells are also shown following that same evolution of frames.
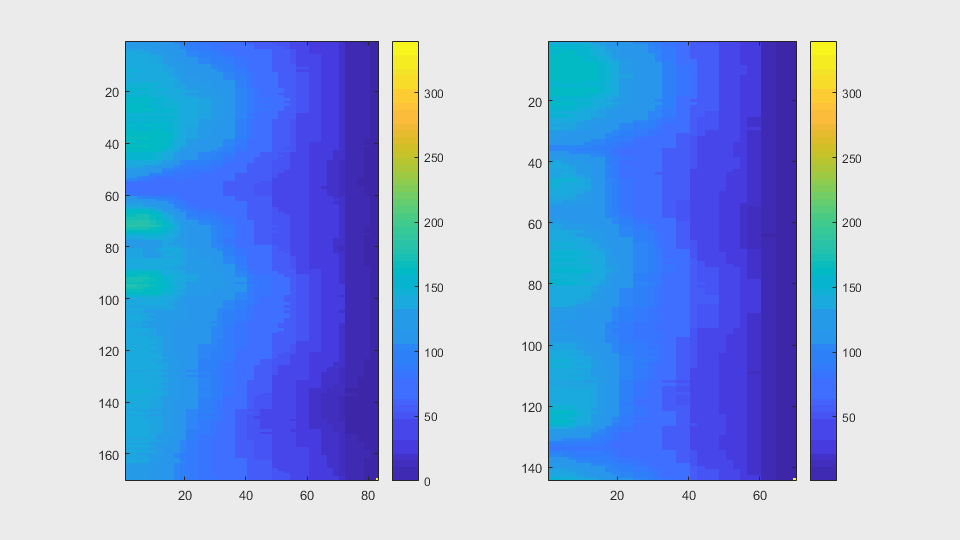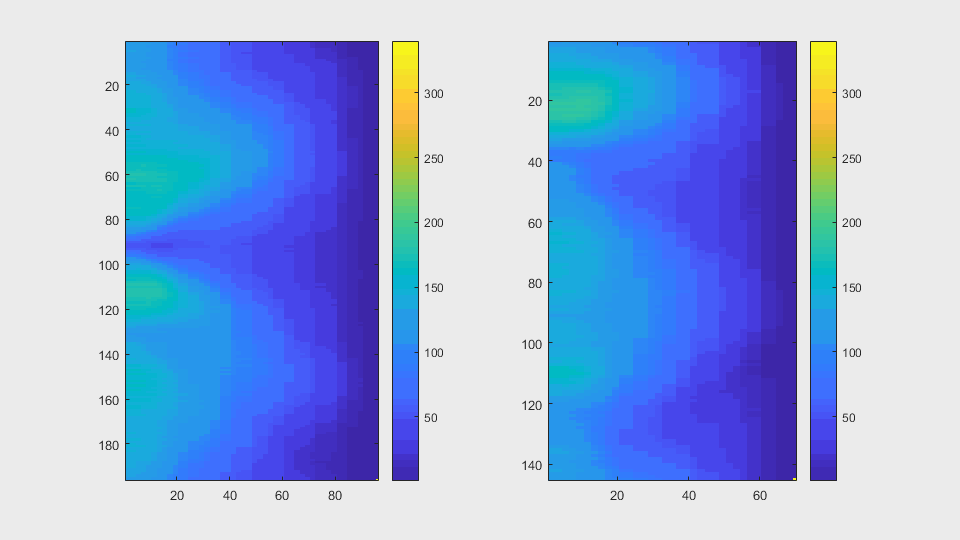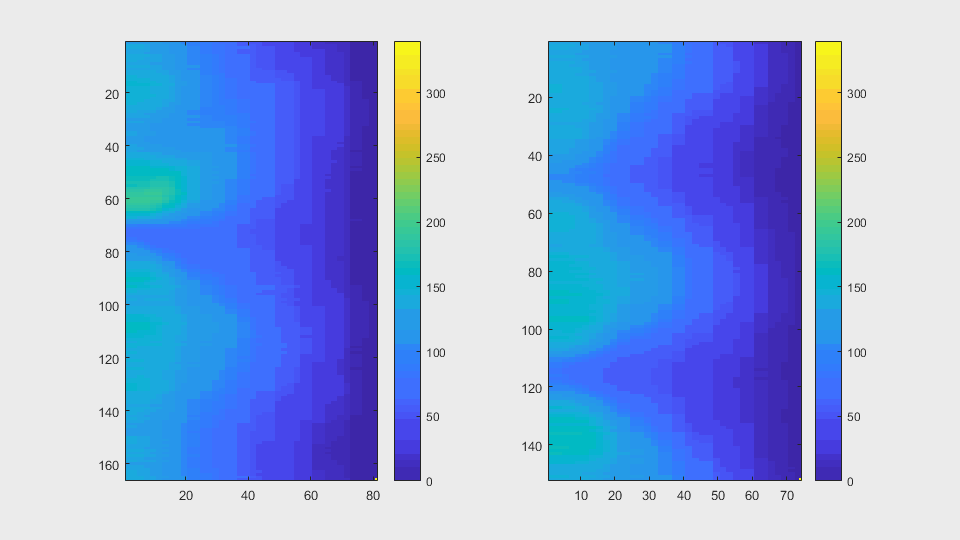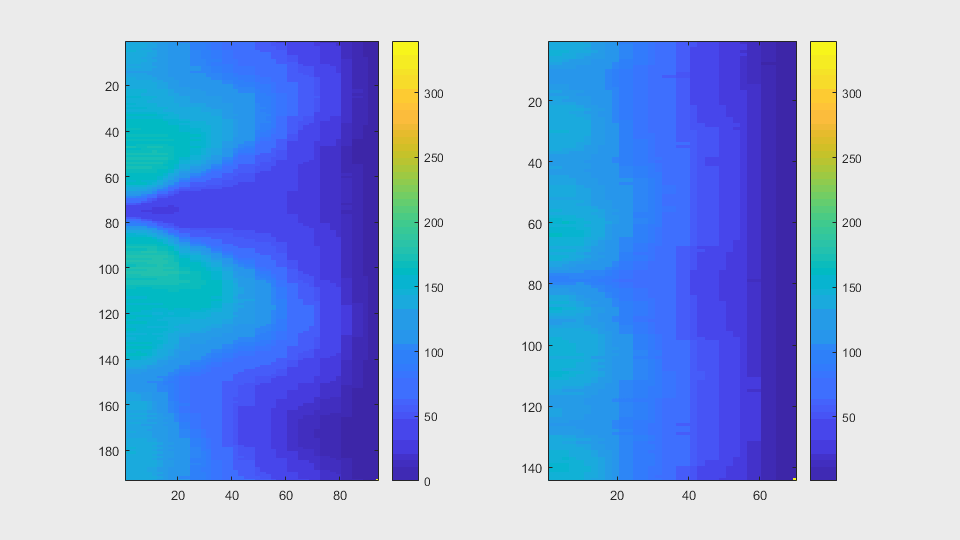
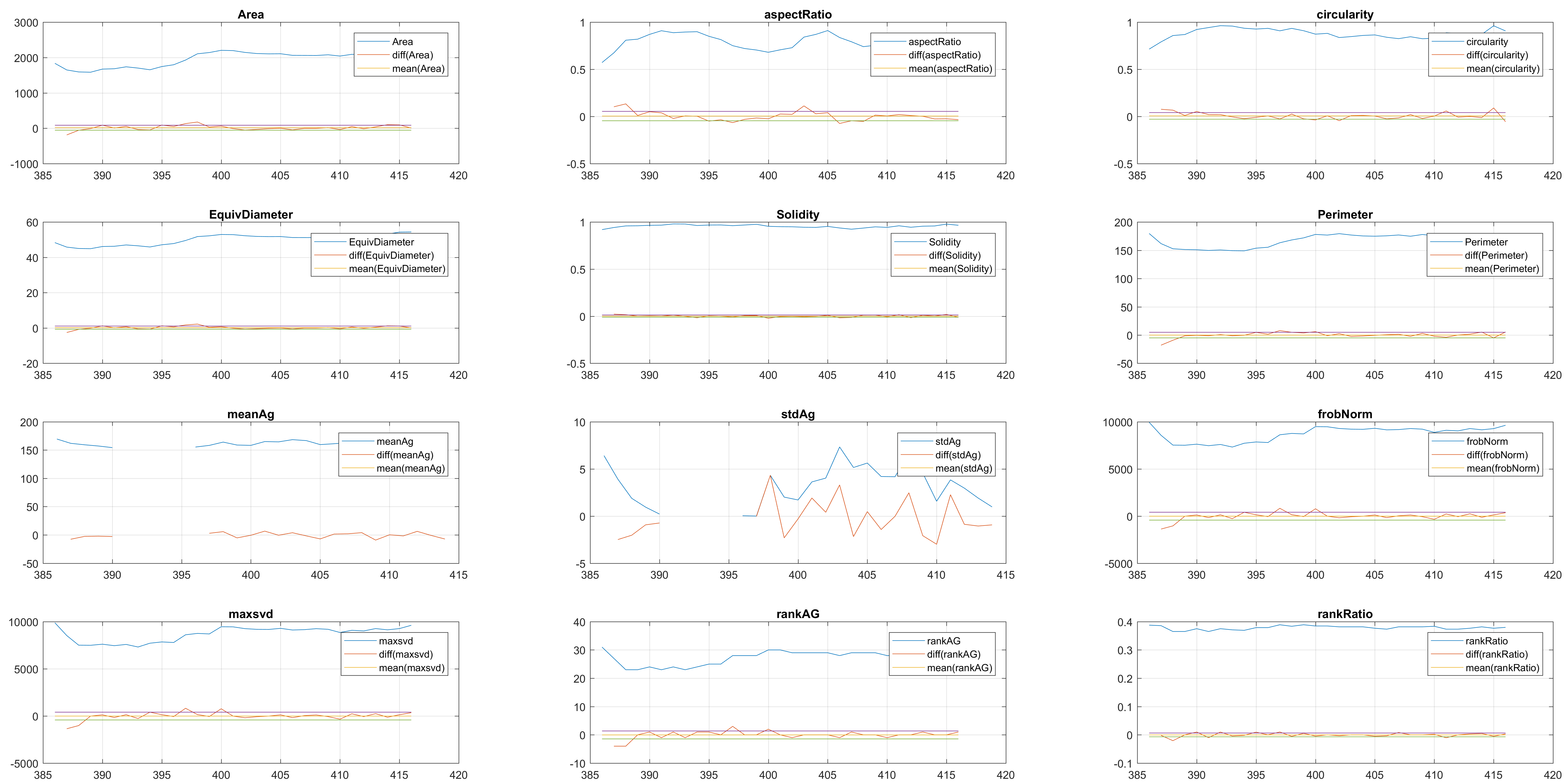
Finally, the measurements with the diff() are shown below.

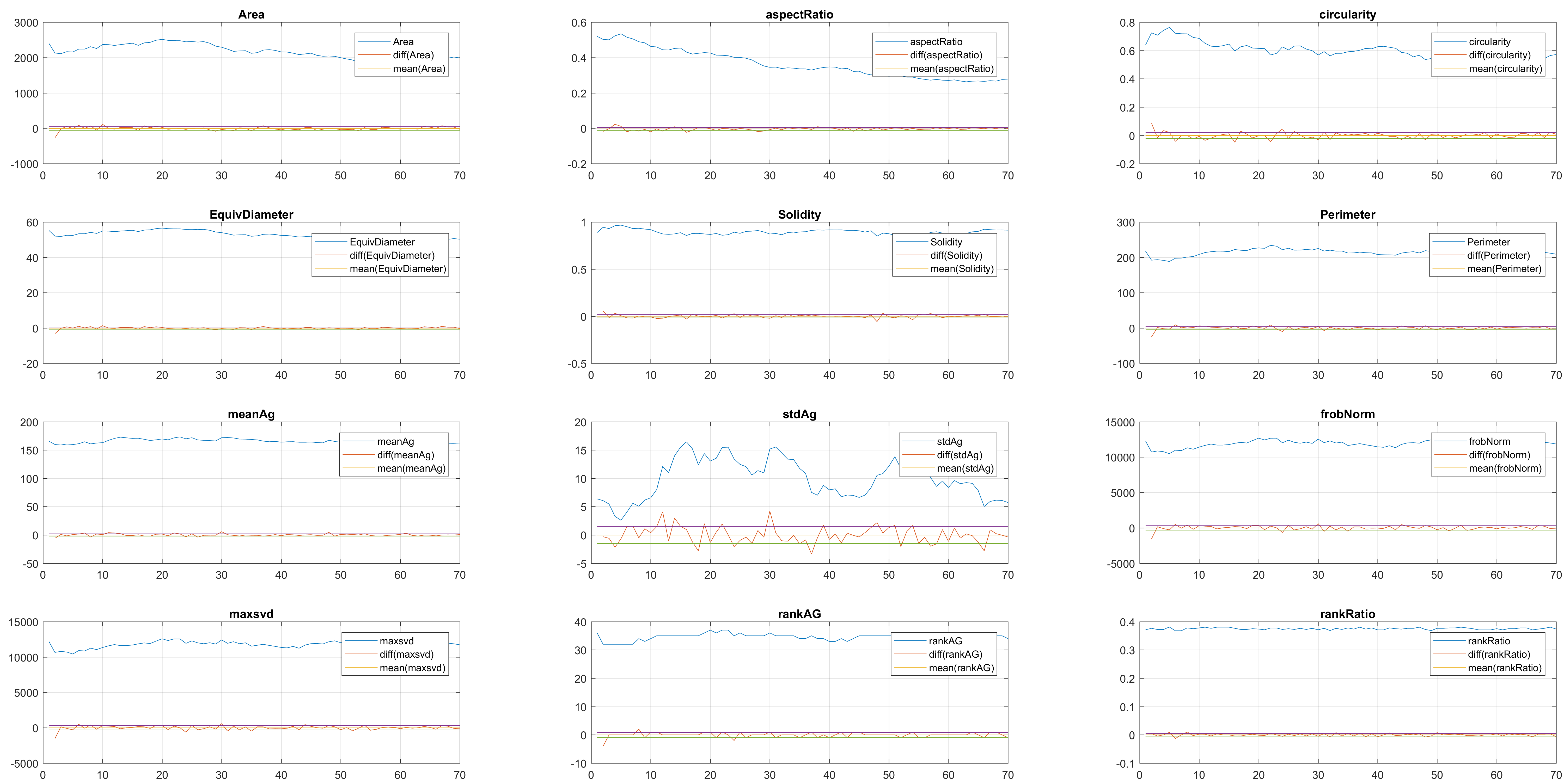
In this case, two non-overlapping cells were followed from wuc=8002 to
assess the different measurements described before. The cells assessed in this
case are as follows:
The anglegrams of both cells are also shown following that same evolution of frames.
Finally, the measurements with the diff() are shown below.
Two overlapping cells were followed from wuc=8007 to
assess the different measurements described before.
Overlapping begins in frame 19.
The cells assessed in this case appear as follows:

The anglegrams of both cells are also shown following that same evolution of frames.
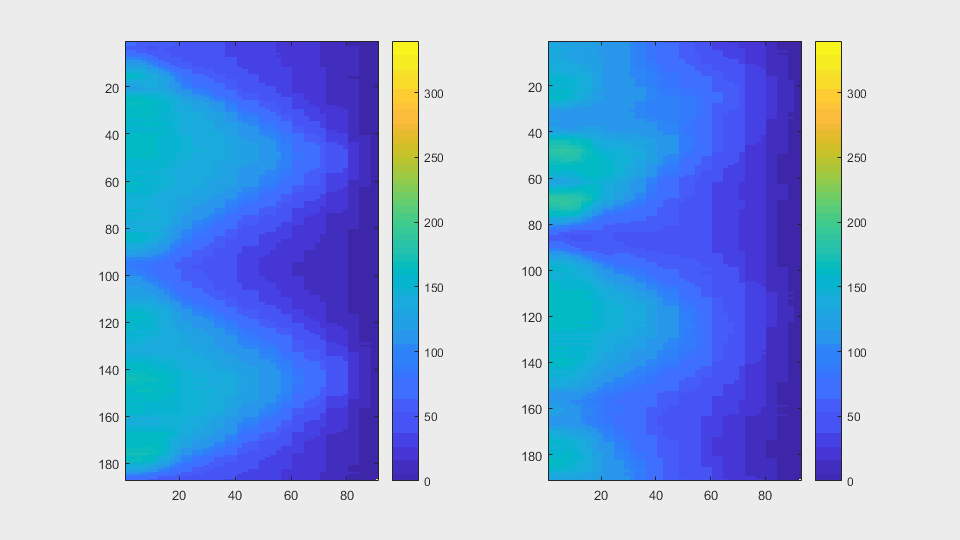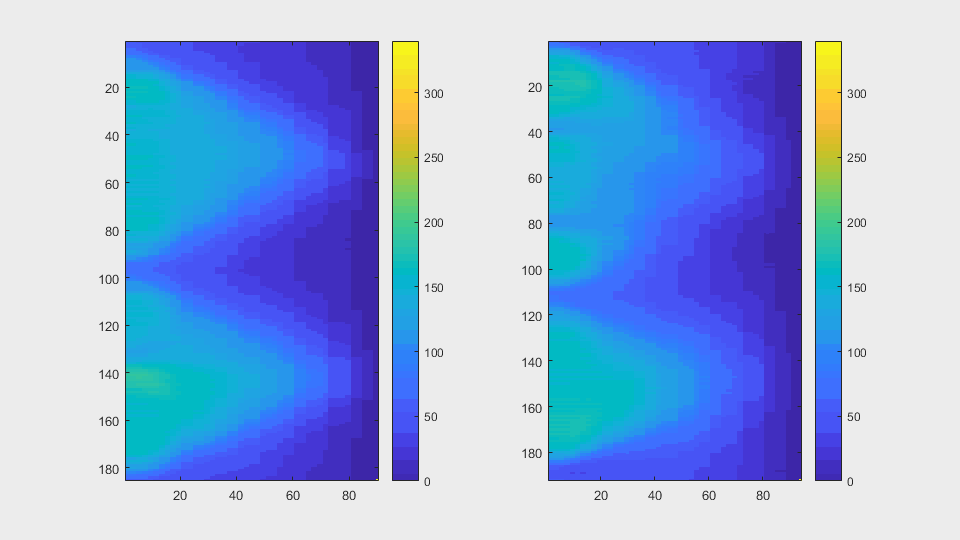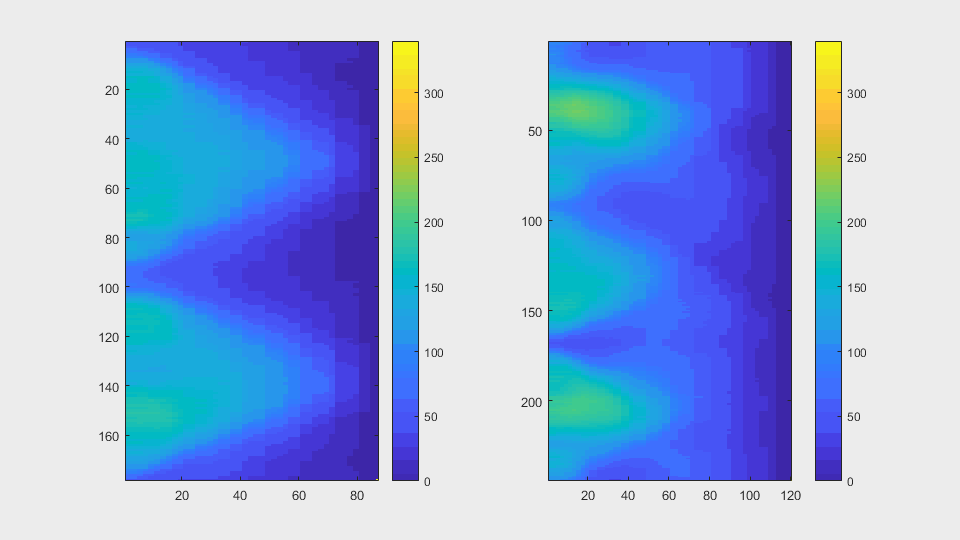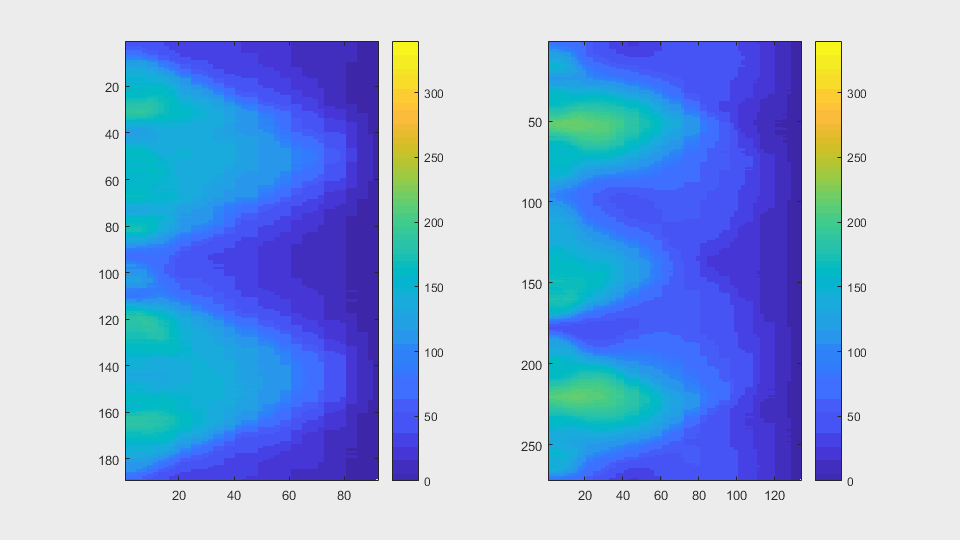
Finally, the measurements with the diff() are shown below.
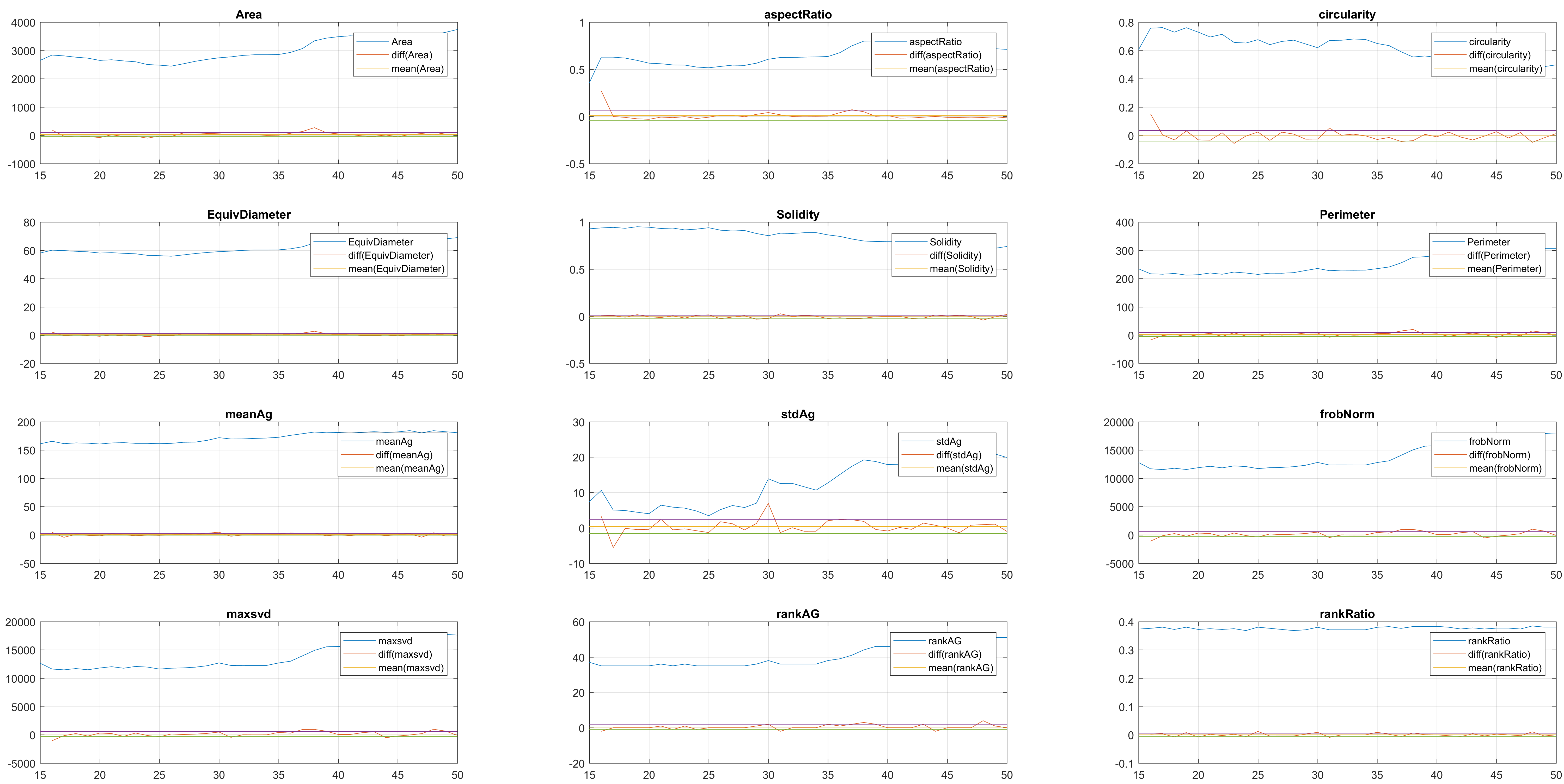
Two overlapping cells were followed from wuc=11010 to
assess the different measurements described before.
Overlapping begins in frame 2.
The cells assessed in this case appear as follows:
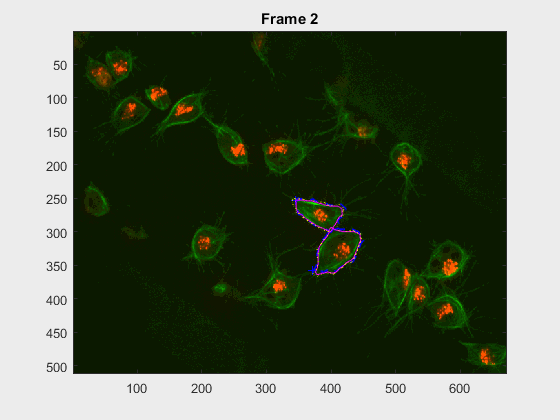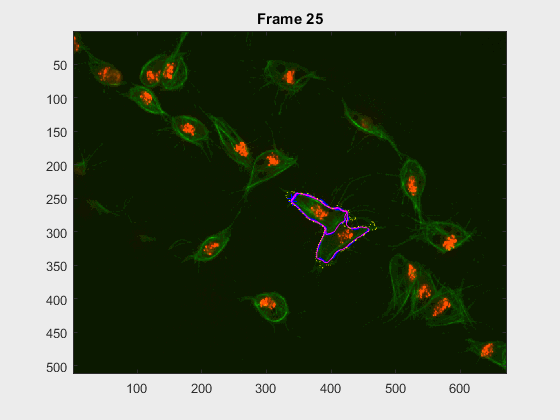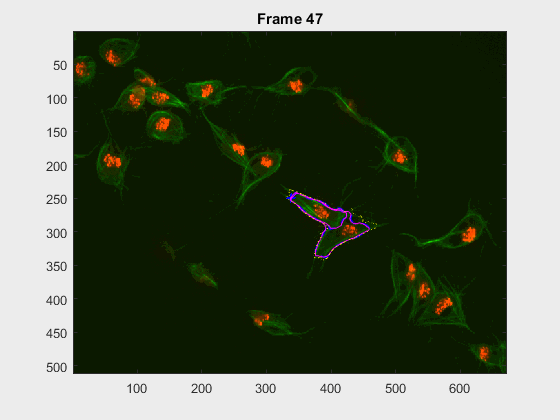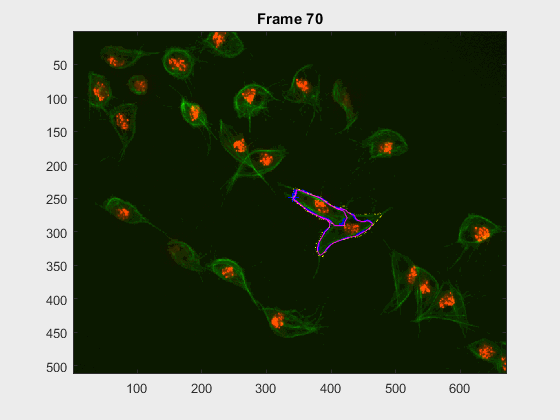
The anglegrams of both cells are also shown following that same evolution of frames.
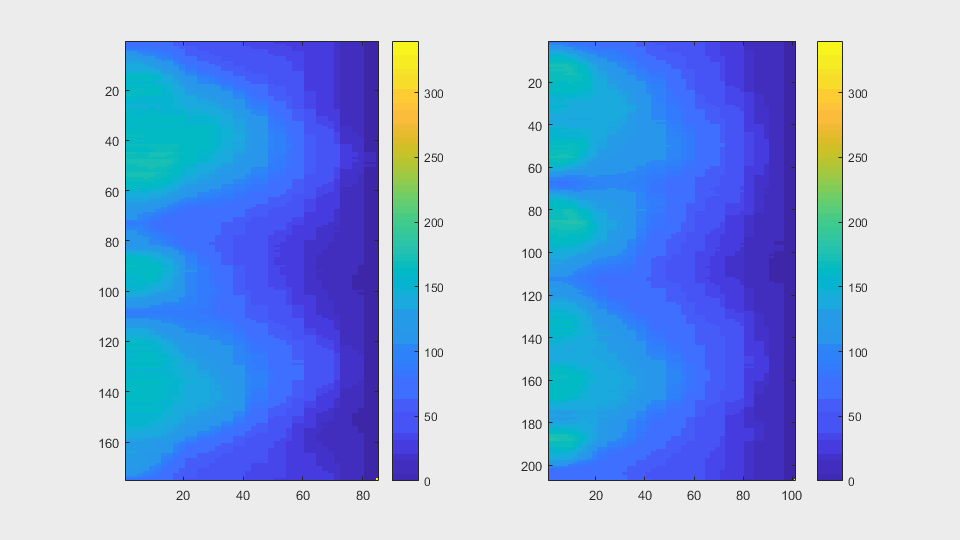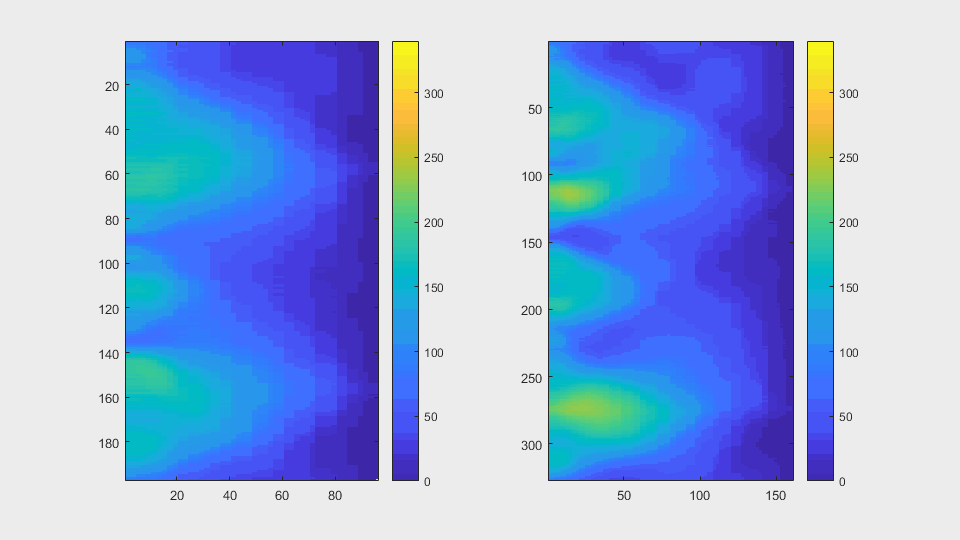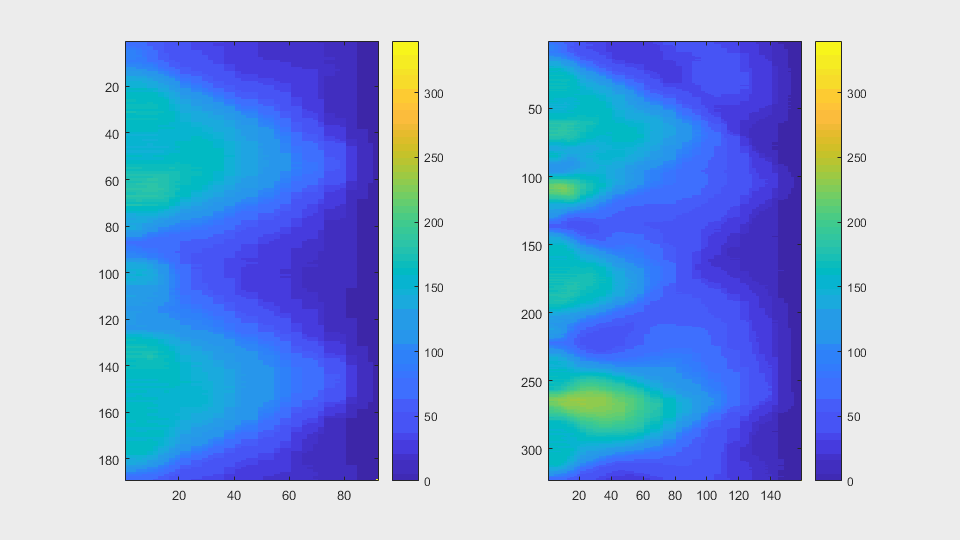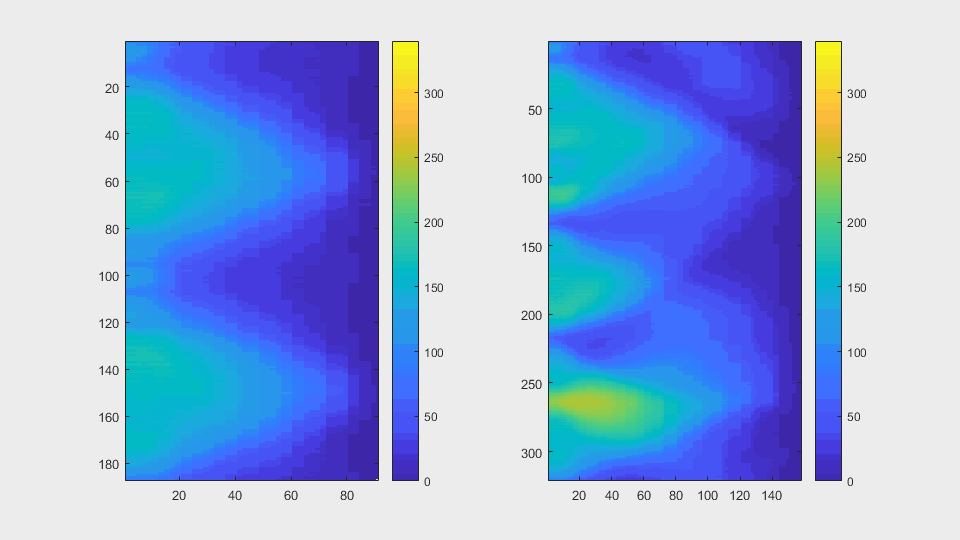
Finally, the measurements with the diff() are shown below.
It can be seen that the cells that appear below, that is cell 8 from
clump wuc=8007 and cell 11 of clump wuc=11010 are the ones where the
segmentation is leaking; whereas the cells that appear above in their
respective clumps have what seems to be a better segmentation.
- (Original idea) use the measurements described here to catch a bad segmentation.
- Since one of the segmentations seems to work... remove that from the frame and try to re-do the bad segmentation...
- Track each point in the boundary as it moves from frame to frame.. many problems, but might work.. have like a keyhole algorithm but for the shape, then weird movements would probably be detected.
The next important step is in this LOG FILE.