diff --git a/DESCRIPTION b/DESCRIPTION
index 72ec297..2fd6f82 100644
--- a/DESCRIPTION
+++ b/DESCRIPTION
@@ -1,19 +1,20 @@
Package: compareDF
Type: Package
Title: Do a Git Style Diff of the Rows Between Two Dataframes with Similar Structure
-Version: 1.8.0
-Date: 2019-06-02
+Version: 2.0.0
+Date: 2020-01-05
Authors@R: person("Alex", "Joseph", email = "alexsanjoseph@gmail.com", role = c("aut", "cre"))
Description: Compares two dataframes which have the same column
structure to show the rows that have changed. Also gives a git style diff format
- to quickly see what has changes in addition to summary statistics.
+ to quickly see what has changed in addition to summary statistics.
License: MIT + file LICENSE
Depends:
- R (>= 3.4.0)
+ R (>= 3.5.0)
Imports:
dplyr (>= 0.4.3),
magrittr (>= 1.5),
htmlTable (>= 1.5),
+ openxlsx (>= 4.1),
tidyr (>= 0.4.1),
stringr (>= 1.0.0)
Suggests:
diff --git a/NAMESPACE b/NAMESPACE
index bfb9739..5ad3337 100644
--- a/NAMESPACE
+++ b/NAMESPACE
@@ -1,6 +1,7 @@
# Generated by roxygen2: do not edit by hand
export(compare_df)
+export(create_output_table)
export(view_html)
import(dplyr)
importFrom(stats,na.omit)
diff --git a/NEWS.md b/NEWS.md
index 3c71d36..d2979e4 100644
--- a/NEWS.md
+++ b/NEWS.md
@@ -1,55 +1,61 @@
-# compareDF 1.8.0
+## compareDF 2.0.0
+* New Major Version! Contains some breaking changes
+* Support for `XLSX` format
+* Write output to file directly
+* Separate functions to compare output and create output tables
+* Cleaner abstractions in functions
+* More bugs squashed
+* Color blind friendly default colors
+
+## compareDF 1.8.0
* Added new option to keep only the columns which have changed using `keep_unchanged_cols`.
* changed option `keep_unchanged` to `keep_unchanged_rows`
-# compareDF 1.7.3
+## compareDF 1.7.3
* Fixed tests to work with dplyr 0.8.2 and on Linux systems
-# compareDF 1.7.2
+## compareDF 1.7.2
* Fixed tests to work with dplyr 0.8.1
-# compareDF 1.7.1
+## compareDF 1.7.1
* Fixed tests to work with dplyr 0.8.0
-# compareDF 1.7.0
+## compareDF 1.7.0
* Provided options to name the columns in the HTML output
* Provided option change column name
* Provided option to change group column name
-# compareDF 1.6.0
+## compareDF 1.6.0
* Added option to specify different types of tolerances. Now you can use `difference` as an argument to use difference rather than ratio
* Fixed some bugs
* Lot more tests
-# compareDF 1.5.0
-* Added an option to preserve the rows that have not changed in the anlysis using the `keep_unchanged_rows` argument
+## compareDF 1.5.0
+* Added an option to preserve the rows that have not changed in the analysis using the `keep_unchanged_rows` argument
* Added an option to set the color scheme in the HTML using the `color_scheme` argument.
* Updated Documentation
* Fixed some bugs
* Fixed dependencies
-# compareDF 1.3.1
+## compareDF 1.3.1
* added a test dependency(stringr) as notified by CRAN
-# compareDF 1.3.0
+## compareDF 1.3.0
* added an option to ignore errors `stop_on_error`
-# compareDF 1.2.0
+## compareDF 1.2.0
* Fixed bugs
* view_html function to view the html file created for setups that cannot print html directly
-# compareDF 1.1.2
+## compareDF 1.1.2
* compareDF can now handle negative inputs correctly
-# compareDF 1.1.1
+## compareDF 1.1.1
* Fixed some bugs when the two frames are similar except reordering
-# compareDF 1.1.0
+## compareDF 1.1.0
* Fixed some bugs
-# compareDF 1.0.0
+## compareDF 1.0.0
* First Release!
-
-
-
diff --git a/R/fnsComparison.R b/R/fnsComparison.R
index a4c62c6..ac0d042 100644
--- a/R/fnsComparison.R
+++ b/R/fnsComparison.R
@@ -8,7 +8,6 @@
#' @param group_col A character vector of a string of character vector showing the columns
#' by which to group_by.
#' @param exclude The columns which should be excluded from the comparison
-#' @param limit_html maximum number of rows to show in the html diff. >1000 not recommended
#' @param stop_on_error Whether to stop on acceptable errors on not
#' @param tolerance The amount in fraction to which changes are ignored while showing the
#' visual representation. By default, the value is 0 and any change in the value of variables
@@ -16,11 +15,6 @@
#' @param tolerance_type Defaults to 'ratio'. The type of comparison for numeric values, can be 'ratio' or 'difference'
#' @param keep_unchanged_rows whether to preserve unchanged values or not. Defaults to \code{FALSE}
#' @param keep_unchanged_cols whether to preserve unchanged values or not. Defaults to \code{TRUE}
-#' @param color_scheme What color scheme to use for the HTML output. Should be a vector/list with
-#' named_elements. Default - \code{c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")}
-#' @param html_headers A character vector of column names to be used in the table. Defaults to \code{colnames}.
-#' @param html_change_col_name Name of the change column to use in the HTML table. Defaults to \code{chng_type}.
-#' @param html_group_col_name Name of the group column to be used in the table (if there are multiple grouping vars). Defaults to \code{grp}.
#' @param round_output_to Number of digits to round the output to. Defaults to 3.
#' @import dplyr
#' @export
@@ -32,10 +26,8 @@
#' ctable = compare_df(new_df, old_df, c("var1"))
#' print(ctable$comparison_df)
#' ctable$html_output
-compare_df <- function(df_new, df_old, group_col, exclude = NULL, limit_html = 100, tolerance = 0, tolerance_type = 'ratio',
+compare_df <- function(df_new, df_old, group_col, exclude = NULL, tolerance = 0, tolerance_type = 'ratio',
stop_on_error = TRUE, keep_unchanged_rows = FALSE, keep_unchanged_cols = TRUE,
- color_scheme = c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue"),
- html_headers = NULL, html_change_col_name = "chng_type", html_group_col_name = "grp",
round_output_to = 3){
both_tables = list(df_new = df_new, df_old = df_old)
@@ -84,20 +76,18 @@ compare_df <- function(df_new, df_old, group_col, exclude = NULL, limit_html = 1
if(nrow(comparison_table) == 0) stop_or_warn("The two data frames are the same after accounting for tolerance!", stop_on_error)
if(nrow(comparison_table_diff) == 0) stop_or_warn("The two data frames are the same after accounting for tolerance!", stop_on_error)
- html_headers_all = get_headers_for_html_table(html_headers, html_change_col_name, html_group_col_name, comparison_table_diff)
-
- if (limit_html > 0 & nrow(comparison_table_diff) > 0 & nrow(comparison_table) > 0)
- html_table = create_html_table(comparison_table_diff, comparison_table_ts2char, group_col, limit_html, color_scheme, html_headers_all) else
- html_table = NULL
change_count = create_change_count(comparison_table, group_col)
change_summary = create_change_summary(change_count, both_tables)
comparison_table$chng_type = comparison_table$chng_type %>% replace_numbers_with_symbols()
- comparison_table_diff = comparison_table_diff %>% replace_numbers_with_symbols()
+ comparison_table_diff_symbols = comparison_table_diff %>% replace_numbers_with_symbols()
- list(comparison_df = comparison_table, html_output = html_table,
- comparison_table_diff = comparison_table_diff,
- change_count = change_count, change_summary = change_summary)
+ list(comparison_df = comparison_table,
+ comparison_table_diff = comparison_table_diff_symbols,
+ change_count = change_count, change_summary = change_summary,
+ group_col = group_col,
+ comparison_table_ts2char = comparison_table_ts2char,
+ comparison_table_diff_numbers = comparison_table_diff)
}
@@ -127,7 +117,9 @@ exclude_columns <- function(both_tables, exclude){
group_columns <- function(both_tables, group_col){
message("Grouping grouping columns")
df_combined = rbind(both_tables$df_new %>% mutate(from = "new"), both_tables$df_old %>% mutate(from = "old"))
- df_combined = df_combined %>% piped.do.call(group_by_, group_col) %>% data.frame(grp = group_indices(.), .) %>% ungroup
+ df_combined = df_combined %>%
+ group_by_at(group_col) %>%
+ data.frame(grp = group_indices(.), .) %>% ungroup()
list(df_new = df_combined %>% filter(from == "new") %>% select(-from),
df_old = df_combined %>% filter(from == "old") %>% select(-from))
}
@@ -155,14 +147,15 @@ create_comparison_table <- function(both_diffs, group_col, round_output_to){
if(nrow(both_diffs$df1_2) != 0) mixed_df = mixed_df %>% rbind(data.frame(chng_type = "1", both_diffs$df1_2))
if(nrow(both_diffs$df2_1) != 0) mixed_df = mixed_df %>% rbind(data.frame(chng_type = "2", both_diffs$df2_1))
mixed_df %>%
- arrange(desc(chng_type)) %>% arrange_(group_col) %>%
+ arrange(desc(chng_type)) %>%
+ arrange_at(group_col) %>%
# mutate(chng_type = ifelse(chng_type == 1, "1", "2")) %>%
select(one_of(group_col), everything()) %>% round_num_cols(round_output_to)
}
create_comparison_table_diff <- function(comparison_table_ts2char, group_col, tolerance, tolerance_type){
- comparison_table_ts2char %>% group_by_(group_col) %>%
+ comparison_table_ts2char %>% group_by_at(group_col) %>%
do(.diff_type_df(., tolerance = tolerance, tolerance_type = tolerance_type)) %>% as.data.frame
}
@@ -172,34 +165,6 @@ eliminate_tolerant_rows <- function(comparison_table, comparison_table_diff){
comparison_table %>% filter(!rows_inside_tolerance)
}
-#' @importFrom utils head
-create_html_table <- function(comparison_table_diff, comparison_table_ts2char, group_col, limit_html, color_scheme, html_headers_all){
-
- comparison_table_ts2char$chng_type = comparison_table_ts2char$chng_type %>% replace_numbers_with_symbols()
-
- if(limit_html > 1000 & comparison_table_diff %>% nrow > 1000)
- warning("Creating HTML diff for a large dataset (>1000 rows) could take a long time!")
-
- if(limit_html < nrow(comparison_table_diff))
- message("Truncating HTML diff table to ", limit_html, " rows...")
-
- requireNamespace("htmlTable")
- comparison_table_color_code = comparison_table_diff %>% do(.colour_coding_df(., color_scheme)) %>% as.data.frame
-
- shading = ifelse(sequence_order_vector(comparison_table_ts2char[[group_col]]) %% 2, "#dedede", "white")
-
- table_css = lapply(comparison_table_color_code, function(x)
- paste0("padding: .2em; color: ", x, ";")) %>% data.frame %>% head(limit_html) %>% as.matrix()
-
- colnames(comparison_table_ts2char) <- html_headers_all
-
- message("Creating HTML table for first ", limit_html, " rows")
- html_table = htmlTable::htmlTable(comparison_table_ts2char %>% head(limit_html),
- col.rgroup = shading,
- rnames = F, css.cell = table_css,
- padding.rgroup = rep("5em", length(shading))
- )
-}
check_if_comparable <- function(df_new, df_old, group_col, stop_on_error){
@@ -223,15 +188,6 @@ round_num_cols <- function(df, round_digits = 2)
df
}
-.colour_coding_df <- function(df, color_scheme){
- if(nrow(df) == 0) return(df)
- df[df == 2] = color_scheme[['addition']]
- df[df == 1] = color_scheme[['removal']]
- df[df == 0] = color_scheme[['unchanged_cell']]
- df[df == -1] = color_scheme[['unchanged_row']]
- df
-}
-
#' @importFrom stats na.omit
.diff_type_df <- function(df, tolerance = 1e-6, tolerance_type = 'ratio'){
@@ -278,8 +234,6 @@ rowdiff <- function(x.1,x.2,...){
df
}
-piped.do.call = function(x, fname, largs) do.call(fname, c(list(x), largs))
-
is.POSIXct <- function(x) inherits(x, "POSIXct")
sequence_order_vector <- function(data)
@@ -289,7 +243,7 @@ sequence_order_vector <- function(data)
}
create_change_count <- function(comparison_table_ts2char, group_col){
- change_count = comparison_table_ts2char %>% group_by_(group_col, "chng_type") %>% tally()
+ change_count = comparison_table_ts2char %>% group_by_at(c(group_col, "chng_type")) %>% tally()
change_count_replace = change_count %>% tidyr::spread(key = chng_type, value = n) %>% data.frame
change_count_replace[is.na(change_count_replace)] = 0
@@ -298,7 +252,7 @@ create_change_count <- function(comparison_table_ts2char, group_col){
change_count_replace = change_count_replace %>% as.data.frame %>%
tidyr::gather_("variable", "value", c("X2", "X1"))
- change_count = change_count_replace %>% group_by_(group_col) %>% arrange_('variable') %>%
+ change_count = change_count_replace %>% group_by_at(group_col) %>% arrange_at('variable') %>%
summarize(changes = min(value), additions = value[2] - value[1], removals = value[1] - value[2]) %>%
mutate(additions = replace(additions, is.na(additions) | additions < 0, 0)) %>%
mutate(removals = replace(removals, is.na(removals) | removals < 0, 0))
@@ -312,7 +266,7 @@ create_change_summary <- function(change_count, both_tables){
changes = sum(change_count$changes), additions = sum(change_count$additions), removals = sum(change_count$removals))
}
-get_headers_for_html_table <- function(headers, change_col_name, group_col_name, comparison_table_diff) {
+get_headers_for_table <- function(headers, change_col_name, group_col_name, comparison_table_diff) {
# if (is.null(headers)) return(names(comparison_table_diff))
headers_all = names(comparison_table_diff) %>%
@@ -324,52 +278,3 @@ get_headers_for_html_table <- function(headers, change_col_name, group_col_name,
headers_all
}
-
-# nocov start
-#' @title View Comparison output HTML
-#'
-#' @description Some versions of Rstudio doesn't automatically show the html pane for the html output. This is a workaround
-#'
-#' @param comparison_output output from the comparisonDF compare function
-#' @export
-#' @examples
-#' old_df = data.frame(var1 = c("A", "B", "C"),
-#' val1 = c(1, 2, 3))
-#' new_df = data.frame(var1 = c("A", "B", "C"),
-#' val1 = c(1, 2, 4))
-#' ctable = compare_df(new_df, old_df, c("var1"))
-#' # Not Run::
-#' # view_html(ctable)
-view_html <- function(comparison_output){
- temp_dir = tempdir()
- temp_file <- paste0(temp_dir, "/temp.html")
- cat(comparison_output$html_output, file = temp_file)
- getOption("viewer")(temp_file)
- unlink("temp.html")
-}
-# nocov end
-
-# Deprecated. Will bring it back in a letter version if deemed necessary
-# create_change_detail_summary <- function(){
-# change_detail = comparison_table_diff
-# change_detail[[group_col]] = comparison_table_ts2char[[group_col]]
-# change_detail = change_detail %>% reshape::melt.data.frame(group_col)
-#
-# change_detail_replace = change_detail %>% group_by_(group_col, "variable", "value") %>% tally()
-# change_detail_replace = change_detail_replace %>% group_by_(group_col, "variable") %>% tidyr::spread(key = value, value = n)
-# change_detail_replace[is.na(change_detail_replace)] = 0
-# change_detail_summary_replace = change_detail_replace %>% data.frame %>% dplyr::rename(param = variable) %>%
-# mutate(param = as.character(param)) %>% tidyr::gather("variable", "value", 3:ncol(.))
-#
-# change_detail_count = change_detail_summary_replace %>% group_by_(group_col, "param") %>% arrange(desc(variable)) %>%
-# summarize(changes = min(value[1:2]), additions = value[1] - value[2], removals = value[2] - value[1]) %>%
-# mutate(additions = replace(additions, is.na(additions), 0)) %>%
-# mutate(removals = replace(removals, is.na(removals), 0))
-# change_detail_count = change_detail_count %>%
-# mutate(replace(changes, changes < 0, 0)) %>%
-# mutate(replace(removals, removals < 0, 0)) %>%
-# mutate(replace(additions, additions < 0, 0))
-#
-# change_detail_count_summary = change_detail_count %>% group_by(param) %>%
-# summarize(total_changes = sum(changes), total_additions = sum(additions), tot_removals = sum(removals))
-# }
diff --git a/R/fnsOutputs.R b/R/fnsOutputs.R
new file mode 100644
index 0000000..c560a51
--- /dev/null
+++ b/R/fnsOutputs.R
@@ -0,0 +1,147 @@
+
+#' @title Create human readable output from the comparison_df output
+#'
+#' @description Currently `html` and `xlsx` are supported
+#'
+#' @param comparison_output Output from the comparison Table functions
+#' @param output_type Type of comparison output. Defaults to `html`
+#' @param file_name Where to write the output to. Default to NULL which output to the Rstudio viewer (not supported for `xlsx`)
+#' @param limit maximum number of rows to show in the diff. >1000 not recommended for HTML
+#' @param color_scheme What color scheme to use for the output. Should be a vector/list with
+#' named_elements. Default - \code{c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")}
+#' @param headers A character vector of column names to be used in the table. Defaults to \code{colnames}.
+#' @param change_col_name Name of the change column to use in the table. Defaults to \code{chng_type}.
+#' @param group_col_name Name of the group column to be used in the table (if there are multiple grouping vars). Defaults to \code{grp}.
+#' @export
+create_output_table <- function(comparison_output, output_type = 'html', file_name = NULL, limit = 100,
+ color_scheme = c("addition" = "#52854C", "removal" = "#FC4E07",
+ "unchanged_cell" = "#999999", "unchanged_row" = "#293352"),
+ headers = NULL, change_col_name = "chng_type", group_col_name = "grp"){
+ headers_all = get_headers_for_table(headers, change_col_name, group_col_name, comparison_output$comparison_table_diff)
+
+ comparison_output$comparison_table_ts2char$chng_type = comparison_output$comparison_table_ts2char$chng_type %>% replace_numbers_with_symbols()
+
+ if (limit == 0 || nrow(comparison_output$comparison_table_diff) == 0 || nrow(comparison_output$comparison_df) == 0)
+ return(NULL)
+ output = switch(output_type,
+ 'html' = create_html_table(comparison_output, file_name, limit, color_scheme, headers_all),
+ 'xlsx' = create_xlsx_document(comparison_output, file_name, limit, color_scheme, headers_all)
+ )
+ output
+}
+
+#' @importFrom utils head
+create_html_table <- function(comparison_output, file_name, limit_html, color_scheme, headers_all){
+
+ comparison_table_diff = comparison_output$comparison_table_diff_numbers
+ comparison_table_ts2char = comparison_output$comparison_table_ts2char
+ group_col = comparison_output$group_col
+
+ if(limit_html > 1000 & comparison_table_diff %>% nrow > 1000)
+ warning("Creating HTML diff for a large dataset (>1000 rows) could take a long time!")
+
+ if(limit_html < nrow(comparison_table_diff))
+ message("Truncating HTML diff table to ", limit_html, " rows...")
+
+ requireNamespace("htmlTable")
+ comparison_table_color_code = comparison_table_diff %>% do(.colour_coding_df(., color_scheme)) %>% as.data.frame
+
+ shading = ifelse(sequence_order_vector(comparison_table_ts2char[[group_col]]) %% 2, "#dedede", "white")
+
+ table_css = lapply(comparison_table_color_code, function(x)
+ paste0("padding: .2em; color: ", x, ";")) %>% data.frame %>% head(limit_html) %>% as.matrix()
+
+ colnames(comparison_table_ts2char) <- headers_all
+
+ message("Creating HTML table for first ", limit_html, " rows")
+ html_table = htmlTable::htmlTable(comparison_table_ts2char %>% head(limit_html),
+ col.rgroup = shading,
+ rnames = F, css.cell = table_css,
+ padding.rgroup = rep("5em", length(shading))
+ )
+ if(!is.null(file_name)){
+ cat(html_table, file = file_name)
+ return(file_name)
+ }
+ return(html_table)
+
+}
+
+.colour_coding_df <- function(df, color_scheme){
+ if(nrow(df) == 0) return(df)
+ df[df == 2] = color_scheme[['addition']]
+ df[df == 1] = color_scheme[['removal']]
+ df[df == 0] = color_scheme[['unchanged_cell']]
+ df[df == -1] = color_scheme[['unchanged_row']]
+ df
+}
+
+.convert_to_row_column_format <- function(x, n){
+ list(
+ rows = ((x - 1) %% n) + 1,
+ cols = ((x - 1) %/% n) + 1
+ )
+}
+
+.get_color_coding_indices <- function(df){
+ output = list(
+ 'addition' = which(df == 2),
+ 'removal' = which(df == 1),
+ 'unchanged_cell' = which(df == 0),
+ 'unchanged_row' = which(df == -1)
+ ) %>% Filter(function(x) length(x) > 0, .) %>%
+ lapply(.convert_to_row_column_format, nrow(df))
+}
+
+create_xlsx_document <- function(comparison_output, file_name, limit, color_scheme, headers_all){
+ if(is.null(file_name)) stop("file_name cannot be null if output format is xlsx")
+ comparison_table_diff = comparison_output$comparison_table_diff_numbers
+ comparison_table_ts2char = comparison_output$comparison_table_ts2char
+ group_col = comparison_output$group_col
+
+ requireNamespace("openxlsx")
+
+ comparison_table_color_code = comparison_table_diff %>% .get_color_coding_indices()
+
+ wb <- openxlsx::createWorkbook("Compare DF Output")
+ openxlsx::addWorksheet(wb, "Sheet1", gridLines = FALSE)
+ openxlsx::writeData(wb, sheet = 1, comparison_table_ts2char, rowNames = FALSE)
+
+ for(i in seq_along(comparison_table_color_code)){
+ openxlsx::addStyle(wb, sheet = 1,
+ openxlsx::createStyle(fontColour = color_scheme[[names(comparison_table_color_code)[i]]]),
+ rows = comparison_table_color_code[[i]]$rows, cols = comparison_table_color_code[[i]]$cols,
+ gridExpand = FALSE)
+ }
+
+ even_rows = which(sequence_order_vector(comparison_table_ts2char[[group_col]]) %% 2 == 0) + 1
+ openxlsx::addStyle(wb, sheet = 1, openxlsx::createStyle(fgFill = 'lightgray'), rows = even_rows,
+ cols = 1:ncol(comparison_table_ts2char), gridExpand = T, stack = TRUE)
+
+ openxlsx::saveWorkbook(wb, file_name, overwrite = T)
+
+}
+
+# nocov start
+#' @title View Comparison output HTML
+#'
+#' @description Some versions of Rstudio doesn't automatically show the html pane for the html output. This is a workaround
+#'
+#' @param comparison_output output from the comparisonDF compare function
+#' @export
+#' @examples
+#' old_df = data.frame(var1 = c("A", "B", "C"),
+#' val1 = c(1, 2, 3))
+#' new_df = data.frame(var1 = c("A", "B", "C"),
+#' val1 = c(1, 2, 4))
+#' ctable = compare_df(new_df, old_df, c("var1"))
+#' # Not Run::
+#' # view_html(ctable)
+view_html <- function(comparison_output){
+ temp_dir = tempdir()
+ temp_file <- paste0(temp_dir, "/temp.html")
+ cat(comparison_output$html_output, file = temp_file)
+ getOption("viewer")(temp_file)
+ unlink("temp.html")
+}
+# nocov end
diff --git a/README.md b/README.md
index 6a784dd..10c2c06 100644
--- a/README.md
+++ b/README.md
@@ -1,9 +1,6 @@
+**New Version released with XLSX Support!!**
-
-**CRAN Version is upto date with the latest bug fixes as of 2019-06-02**
-
-
-# compareDF
+## compareDF
[](https://codecov.io/github/alexsanjoseph/compareDF?branch=master)
[](https://travis-ci.org/alexsanjoseph/compareDF)
@@ -13,7 +10,7 @@


-# Introduction
+## Introduction
Every so often while doing data analysis, I have come across a situation where I have
two datasets, which have the same structure but with small differences in the actual
@@ -29,14 +26,16 @@ the data frame and lesser on the data itself. I was not able to easily identify
isolate what has changed in the data itself. So I decided to write one for myself. That is
what `compareDF` package is all about.
-# Usage
+The output can be visualized either on the RStudio Viewer, or sent to a file as an `HTML` or an `XLSX` file.
+
+## Usage
The package has a single function, `compare_df`. It takes in two data frames, and one or
more grouping variables and does a comparison between the the two. In addition you can
specify columns to ignore, decide how many rows of changes to be displayed in the case
of the HTML output, and decide what tolerance you want to provide to detect change.
-# Basic Example
+## Basic Example
Let's take the example of a teacher who wants to compare the marks and grades of students across
two years, 2010 and 2011. The data is stored in tabular format.
@@ -59,7 +58,8 @@ Eg: - _Mugger and Dhakkan_ dropped out while _Vikram and Dikchik_ where added in
The package allows a user to quickly identify these changes.
-## Basic Comparison
+### Basic Comparison
+
Now let's compare the performance of the students across the years. The grouping variables are the
_Student_ column. We will ignore the _Division_ column and assume that the student names are unique
across divisions. In this sub-example, if a student appears in two divisions, he/she has studied in both
@@ -84,14 +84,14 @@ the two years so that row is included.
However, _Macho, Division B_ has had the exact same scores in both the years for all subjects, so his data is not
shown in the comparison table.
-## HTML Output
+### HTML Output
While the comparison table can be quickly summarized in various forms for futher analysis, it is
very difficult to process visually. The `html_output` provides a way to represent this is a way that is easier
for the numan eye to read. NOTE: You need to install the `htmlTable` package for the HTML comparison to work.
_For the purpose of the readme I am attaching the html as a png because github markdown doesn't retain styles._
```{r, results = 'hide'}
-print(ctable_student$html_output)
+create_output_table(ctable_student)
```
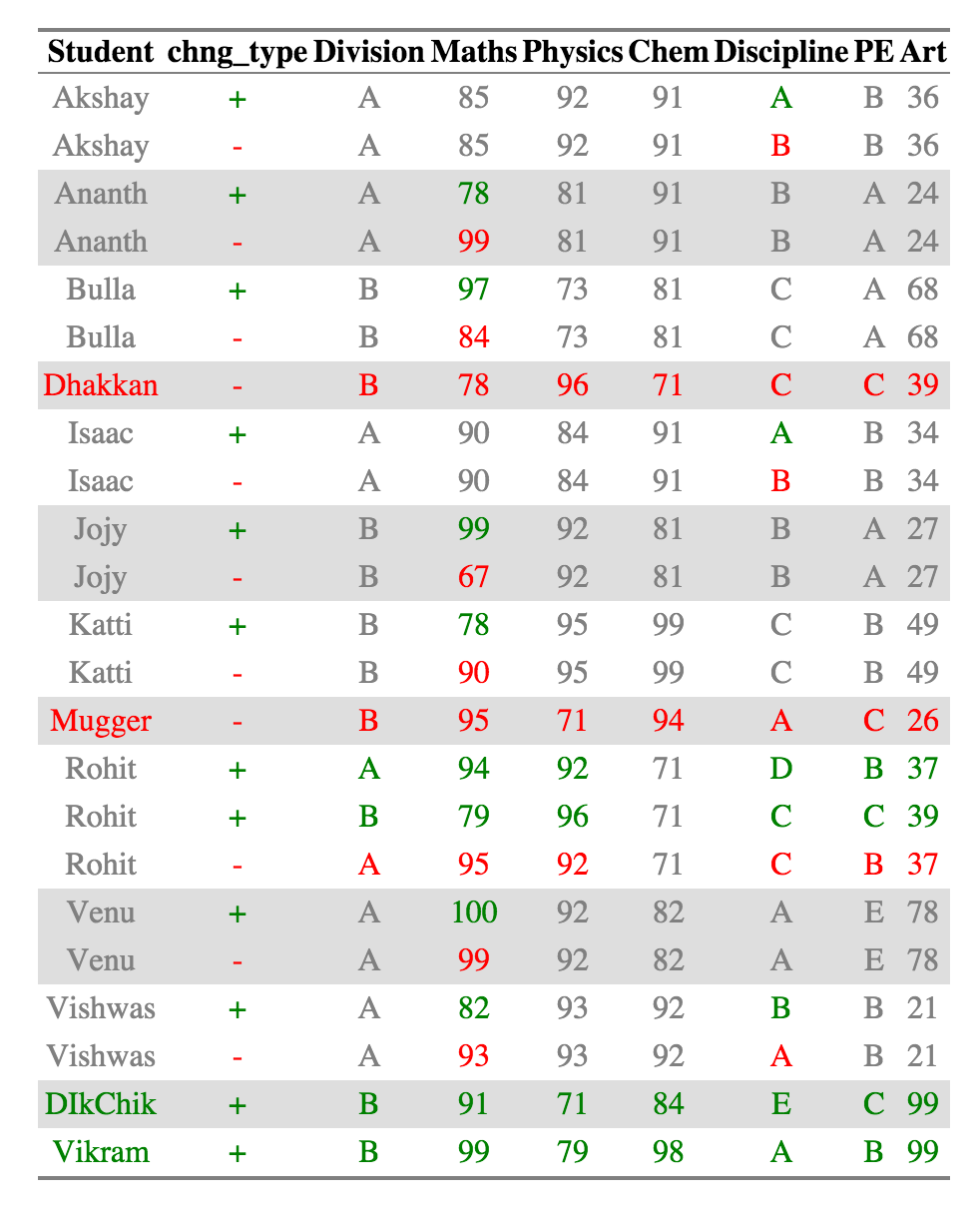 @@ -109,11 +109,16 @@ scores, which are in _Discipline_, _Maths_, and _Maths_ respectively.
_Dhakkan_ and _Mugger_ have dropped out of the dataset from 2010 and the all the columns for the rows are shown
in red, which _DikChik_ and _Vikram_ have joined new in the data set and all the columns for the rows are in green.
-The same data is represented in tabular form (for further analysis, if necessary) in the
-`comparison_table_diff` object
+```{r}
+create_output_table(ctable_student)
+```
+
+### XLSX Output
+
+Alternately you can write to an xlsx file as well
```{r}
-ctable_student$comparison_table_diff
+create_output_table(ctable_student, output_type = 'xlsx', file_name = "test_file.xlsx")
```
## Change Count and Summary
@@ -129,7 +134,7 @@ ctable_student$change_count
ctable_student$change_summary
```
-## Grouping Multiple Columns
+### Grouping Multiple Columns
We can also group_multiple columns into the grouping variable
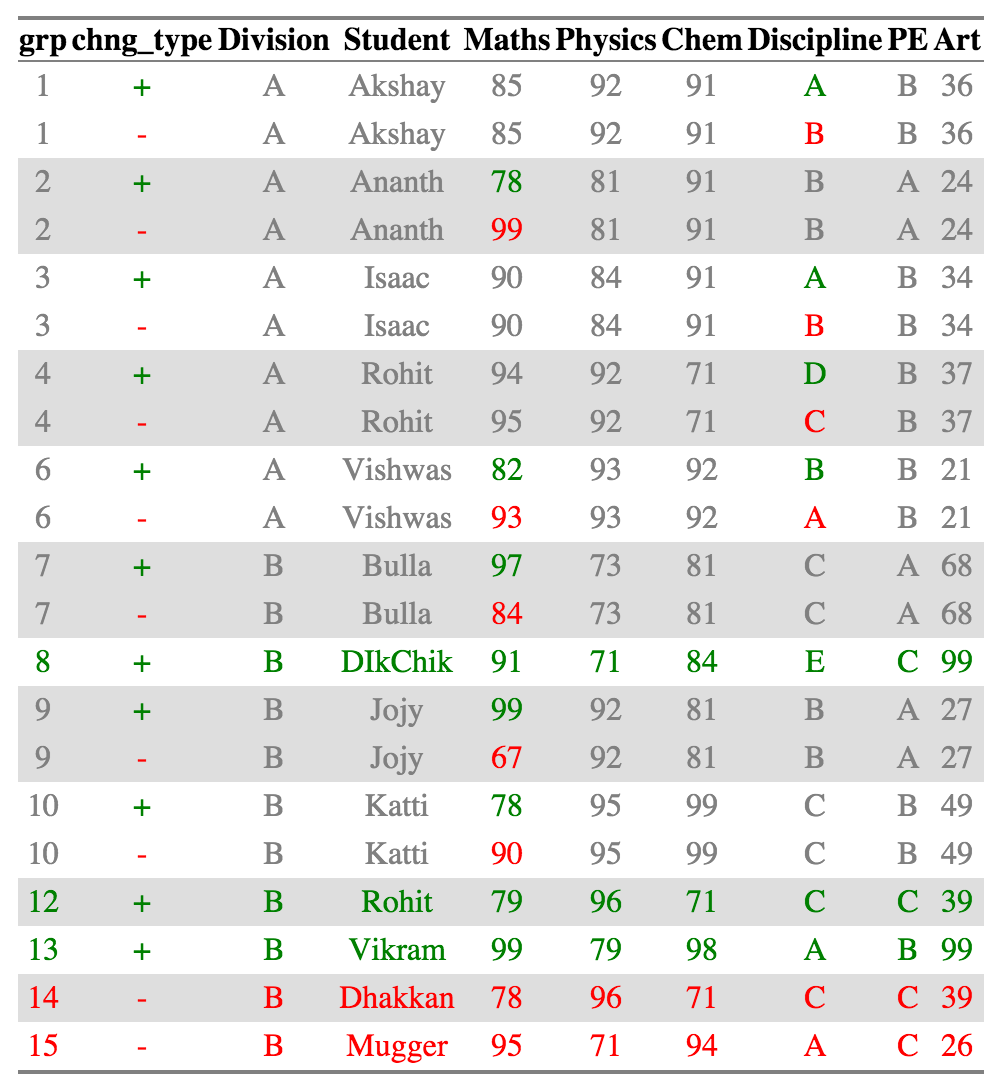
@@ -142,18 +147,17 @@ ctable_student_div$comparison_df
Now _Rohits_ in each individual division are considered as belonging to separate
groups and compared accordingly. All the other summaries also change appropriately.
-## Excluding certain Columns
+### Excluding certain Columns
-You can ignore certain columns using the *exclude* parameter. The fields that have to be
+You can ignore certain columns using the _exclude_ parameter. The fields that have to be
excluded can be given as a character vector. (This is a convenience function to deal with
the case where some columns are not included)
-## Preserving all rows
+### Preserving all rows
The default behavior of the `compare_df` function is to show only the records that have changed. Sometimes the use might want to preserve all the records even after the comparison and not just see the values that have been changed (especially in the HTML). In this case, you can set the `keep_unchanged` parameter to `TRUE`.
-
-## Limiting HTML size
+### Limiting HTML size
For dataframes which have a large amount of differences in them, generating HTML might take
a long time and crash your system. So the maximum diff size for the HTML (and for the HTML
@@ -161,12 +165,11 @@ visualization only) is capped at 100 by default. If you want to see more differe
the `limit_html` parameter appropriately. NOTE: This is only of the HTML output which is used for visual
checking. The main comparison data frame and the summaries ALWAYS include data from all the rows.
-## Changing HTML color
-
-You can use the `color_scheme` parameter to change the color of the HTML generated. The parameter must me a named vector or a list with the appropriate names - The default values are `c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")` but can be changed as needed by the user.
+### Changing color
+You can use the `color_scheme` parameter to change the color of the cells generated. The parameter must me a named vector or a list with the appropriate names - The default values are `c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")` but can be changed as needed by the user.
-## Tolerance
+### Tolerance
It is possible that you'd like numbers very close to each other to be ignored. For example,
if the marks of a student vary by less that 5% across the years, it could be due to random
@@ -176,7 +179,7 @@ less than 5% apart from the lower value.
```{r, results = 'hide'}
ctable_student_div = compare_df(results_2011, results_2010, c("Division", "Student"), tolerance = 0.05)
-ctable_student_div$html_output
+create_output_table(ctable_student_div)
```
@@ -109,11 +109,16 @@ scores, which are in _Discipline_, _Maths_, and _Maths_ respectively.
_Dhakkan_ and _Mugger_ have dropped out of the dataset from 2010 and the all the columns for the rows are shown
in red, which _DikChik_ and _Vikram_ have joined new in the data set and all the columns for the rows are in green.
-The same data is represented in tabular form (for further analysis, if necessary) in the
-`comparison_table_diff` object
+```{r}
+create_output_table(ctable_student)
+```
+
+### XLSX Output
+
+Alternately you can write to an xlsx file as well
```{r}
-ctable_student$comparison_table_diff
+create_output_table(ctable_student, output_type = 'xlsx', file_name = "test_file.xlsx")
```
## Change Count and Summary
@@ -129,7 +134,7 @@ ctable_student$change_count
ctable_student$change_summary
```
-## Grouping Multiple Columns
+### Grouping Multiple Columns
We can also group_multiple columns into the grouping variable
@@ -142,18 +147,17 @@ ctable_student_div$comparison_df
Now _Rohits_ in each individual division are considered as belonging to separate
groups and compared accordingly. All the other summaries also change appropriately.
-## Excluding certain Columns
+### Excluding certain Columns
-You can ignore certain columns using the *exclude* parameter. The fields that have to be
+You can ignore certain columns using the _exclude_ parameter. The fields that have to be
excluded can be given as a character vector. (This is a convenience function to deal with
the case where some columns are not included)
-## Preserving all rows
+### Preserving all rows
The default behavior of the `compare_df` function is to show only the records that have changed. Sometimes the use might want to preserve all the records even after the comparison and not just see the values that have been changed (especially in the HTML). In this case, you can set the `keep_unchanged` parameter to `TRUE`.
-
-## Limiting HTML size
+### Limiting HTML size
For dataframes which have a large amount of differences in them, generating HTML might take
a long time and crash your system. So the maximum diff size for the HTML (and for the HTML
@@ -161,12 +165,11 @@ visualization only) is capped at 100 by default. If you want to see more differe
the `limit_html` parameter appropriately. NOTE: This is only of the HTML output which is used for visual
checking. The main comparison data frame and the summaries ALWAYS include data from all the rows.
-## Changing HTML color
-
-You can use the `color_scheme` parameter to change the color of the HTML generated. The parameter must me a named vector or a list with the appropriate names - The default values are `c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")` but can be changed as needed by the user.
+### Changing color
+You can use the `color_scheme` parameter to change the color of the cells generated. The parameter must me a named vector or a list with the appropriate names - The default values are `c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")` but can be changed as needed by the user.
-## Tolerance
+### Tolerance
It is possible that you'd like numbers very close to each other to be ignored. For example,
if the marks of a student vary by less that 5% across the years, it could be due to random
@@ -176,7 +179,7 @@ less than 5% apart from the lower value.
```{r, results = 'hide'}
ctable_student_div = compare_df(results_2011, results_2010, c("Division", "Student"), tolerance = 0.05)
-ctable_student_div$html_output
+create_output_table(ctable_student_div)
```
 @@ -186,13 +189,13 @@ diff calculation or in the output
Naturally, tolerance has no meaning for non-numeric values.
## Additional features
-- set the color scheme in the HTML using the `color_scheme` argument
-- preserve the rows that have not changed in the anlysis using the `keep_unchanged_rows` argument
-- use `difference` as an argument to use difference rather than ratio for tolerance
-- options to name the columns in the HTML output
-- option change column name
-- option to change group column name
-- keep only the columns which have changed using `keep_unchanged_cols`
+* set the color scheme in the HTML using the `color_scheme` argument
+* preserve the rows that have not changed in the analysis using the `keep_unchanged_rows` argument
+* use `difference` as an argument to use difference rather than ratio for tolerance
+* options to name the columns in the HTML output
+* option change column name
+* option to change group column name
+* keep only the columns which have changed using `keep_unchanged_cols`
## Using compare DF in GAP analysis
@@ -203,8 +206,8 @@ The compareDF package can be used to conduct effective Gap analyses. If the pack
Thanks to Nitin for proofreading the doc and making sure everything made sense.
## Contributors
-- Brice Richard
-- Joshua David Barillas - https://github.com/jdbarillas
+* Brice Richard
+* Joshua David Barillas -
License (MIT)
diff --git a/cran-comments.md b/cran-comments.md
index 4f8e67c..ff211ff 100644
--- a/cran-comments.md
+++ b/cran-comments.md
@@ -1,14 +1,22 @@
## Resubmission
This is a new version. In this version I have:
-* Added new option to keep only the columns which have changed.
-* changed option `keep_unchanged` to `keep_unchanged_rows`
+* Made breaking changes to the package, hence a new major version number
+* Support for `XLSX` format
+* Write output to file directly
+* Separate functions to compare output and create output tables
+* Cleaner abstractions in functions
+* More bugs squashed
+* Color blind friendly default colors
## Test environments
-* local OS X install - 10.14, R 3.5.1
-* ubuntu 16.04 (on travis-ci)
-* win-builder (devel and release)
-* Windows (on Appveyor)
+* local OS X install - 10.14, R 3.5.1
+* ubuntu 16.04 (on travis-ci)
+* win-builder (devel and release)
+* Windows (on Appveyor)
+* Windows Server 2008 R2 SP1, R-devel, 32/64 bit (Rhub)
+* Ubuntu Linux 16.04 LTS, R-release, GCC (Rhub)
+* Fedora Linux, R-devel, clang, gfortran (Rhub)
## R CMD check results
@@ -18,4 +26,3 @@ This is a new version. In this version I have:
There are no reverse dependencies.
-
diff --git a/man/compare_df.Rd b/man/compare_df.Rd
index 7899d5b..e9f844c 100644
--- a/man/compare_df.Rd
+++ b/man/compare_df.Rd
@@ -4,12 +4,9 @@
\alias{compare_df}
\title{Compare Two dataframes}
\usage{
-compare_df(df_new, df_old, group_col, exclude = NULL, limit_html = 100,
- tolerance = 0, tolerance_type = "ratio", stop_on_error = TRUE,
+compare_df(df_new, df_old, group_col, exclude = NULL, tolerance = 0,
+ tolerance_type = "ratio", stop_on_error = TRUE,
keep_unchanged_rows = FALSE, keep_unchanged_cols = TRUE,
- color_scheme = c(addition = "green", removal = "red", unchanged_cell =
- "gray", unchanged_row = "deepskyblue"), html_headers = NULL,
- html_change_col_name = "chng_type", html_group_col_name = "grp",
round_output_to = 3)
}
\arguments{
@@ -22,8 +19,6 @@ by which to group_by.}
\item{exclude}{The columns which should be excluded from the comparison}
-\item{limit_html}{maximum number of rows to show in the html diff. >1000 not recommended}
-
\item{tolerance}{The amount in fraction to which changes are ignored while showing the
visual representation. By default, the value is 0 and any change in the value of variables
is shown off. Doesn't apply to categorical variables.}
@@ -36,15 +31,6 @@ is shown off. Doesn't apply to categorical variables.}
\item{keep_unchanged_cols}{whether to preserve unchanged values or not. Defaults to \code{TRUE}}
-\item{color_scheme}{What color scheme to use for the HTML output. Should be a vector/list with
-named_elements. Default - \code{c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")}}
-
-\item{html_headers}{A character vector of column names to be used in the table. Defaults to \code{colnames}.}
-
-\item{html_change_col_name}{Name of the change column to use in the HTML table. Defaults to \code{chng_type}.}
-
-\item{html_group_col_name}{Name of the group column to be used in the table (if there are multiple grouping vars). Defaults to \code{grp}.}
-
\item{round_output_to}{Number of digits to round the output to. Defaults to 3.}
}
\description{
diff --git a/man/create_output_table.Rd b/man/create_output_table.Rd
new file mode 100644
index 0000000..b4229ce
--- /dev/null
+++ b/man/create_output_table.Rd
@@ -0,0 +1,33 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/fnsOutputs.R
+\name{create_output_table}
+\alias{create_output_table}
+\title{Create human readable output from the comparison_df output}
+\usage{
+create_output_table(comparison_output, output_type = "html",
+ file_name = NULL, limit = 100, color_scheme = c(addition =
+ "#52854C", removal = "#FC4E07", unchanged_cell = "#999999", unchanged_row
+ = "#293352"), headers = NULL, change_col_name = "chng_type",
+ group_col_name = "grp")
+}
+\arguments{
+\item{comparison_output}{Output from the comparison Table functions}
+
+\item{output_type}{Type of comparison output. Defaults to `html`}
+
+\item{file_name}{Where to write the output to. Default to NULL which output to the Rstudio viewer (not supported for `xlsx`)}
+
+\item{limit}{maximum number of rows to show in the diff. >1000 not recommended for HTML}
+
+\item{color_scheme}{What color scheme to use for the output. Should be a vector/list with
+named_elements. Default - \code{c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")}}
+
+\item{headers}{A character vector of column names to be used in the table. Defaults to \code{colnames}.}
+
+\item{change_col_name}{Name of the change column to use in the table. Defaults to \code{chng_type}.}
+
+\item{group_col_name}{Name of the group column to be used in the table (if there are multiple grouping vars). Defaults to \code{grp}.}
+}
+\description{
+Currently `html` and `xlsx` are supported
+}
diff --git a/man/view_html.Rd b/man/view_html.Rd
index 43724b6..17313e1 100644
--- a/man/view_html.Rd
+++ b/man/view_html.Rd
@@ -1,5 +1,5 @@
% Generated by roxygen2: do not edit by hand
-% Please edit documentation in R/fnsComparison.R
+% Please edit documentation in R/fnsOutputs.R
\name{view_html}
\alias{view_html}
\title{View Comparison output HTML}
diff --git a/tests/testthat/test-fnsComparison.R b/tests/testthat/test-fnsComparison.R
index e2f1ad4..f8193fd 100644
--- a/tests/testthat/test-fnsComparison.R
+++ b/tests/testthat/test-fnsComparison.R
@@ -150,16 +150,9 @@ expected_comparison_df = data.frame(grp = c(3, 4),
# arrange(desc(chng_type)) %>% arrange_("var1")
expect_equal(ctable$comparison_df, expected_comparison_df)
-#===============================================================================
-# Limit
-context("compare_df: limit")
-max_rows = 2
-ctable = compare_df(new_df, old_df, c("var1", "var2"), limit_html = max_rows)
-expect_equal(ctable$html_output %>% as.character() %>% stringr::str_count("
@@ -186,13 +189,13 @@ diff calculation or in the output
Naturally, tolerance has no meaning for non-numeric values.
## Additional features
-- set the color scheme in the HTML using the `color_scheme` argument
-- preserve the rows that have not changed in the anlysis using the `keep_unchanged_rows` argument
-- use `difference` as an argument to use difference rather than ratio for tolerance
-- options to name the columns in the HTML output
-- option change column name
-- option to change group column name
-- keep only the columns which have changed using `keep_unchanged_cols`
+* set the color scheme in the HTML using the `color_scheme` argument
+* preserve the rows that have not changed in the analysis using the `keep_unchanged_rows` argument
+* use `difference` as an argument to use difference rather than ratio for tolerance
+* options to name the columns in the HTML output
+* option change column name
+* option to change group column name
+* keep only the columns which have changed using `keep_unchanged_cols`
## Using compare DF in GAP analysis
@@ -203,8 +206,8 @@ The compareDF package can be used to conduct effective Gap analyses. If the pack
Thanks to Nitin for proofreading the doc and making sure everything made sense.
## Contributors
-- Brice Richard
-- Joshua David Barillas - https://github.com/jdbarillas
+* Brice Richard
+* Joshua David Barillas -
License (MIT)
diff --git a/cran-comments.md b/cran-comments.md
index 4f8e67c..ff211ff 100644
--- a/cran-comments.md
+++ b/cran-comments.md
@@ -1,14 +1,22 @@
## Resubmission
This is a new version. In this version I have:
-* Added new option to keep only the columns which have changed.
-* changed option `keep_unchanged` to `keep_unchanged_rows`
+* Made breaking changes to the package, hence a new major version number
+* Support for `XLSX` format
+* Write output to file directly
+* Separate functions to compare output and create output tables
+* Cleaner abstractions in functions
+* More bugs squashed
+* Color blind friendly default colors
## Test environments
-* local OS X install - 10.14, R 3.5.1
-* ubuntu 16.04 (on travis-ci)
-* win-builder (devel and release)
-* Windows (on Appveyor)
+* local OS X install - 10.14, R 3.5.1
+* ubuntu 16.04 (on travis-ci)
+* win-builder (devel and release)
+* Windows (on Appveyor)
+* Windows Server 2008 R2 SP1, R-devel, 32/64 bit (Rhub)
+* Ubuntu Linux 16.04 LTS, R-release, GCC (Rhub)
+* Fedora Linux, R-devel, clang, gfortran (Rhub)
## R CMD check results
@@ -18,4 +26,3 @@ This is a new version. In this version I have:
There are no reverse dependencies.
-
diff --git a/man/compare_df.Rd b/man/compare_df.Rd
index 7899d5b..e9f844c 100644
--- a/man/compare_df.Rd
+++ b/man/compare_df.Rd
@@ -4,12 +4,9 @@
\alias{compare_df}
\title{Compare Two dataframes}
\usage{
-compare_df(df_new, df_old, group_col, exclude = NULL, limit_html = 100,
- tolerance = 0, tolerance_type = "ratio", stop_on_error = TRUE,
+compare_df(df_new, df_old, group_col, exclude = NULL, tolerance = 0,
+ tolerance_type = "ratio", stop_on_error = TRUE,
keep_unchanged_rows = FALSE, keep_unchanged_cols = TRUE,
- color_scheme = c(addition = "green", removal = "red", unchanged_cell =
- "gray", unchanged_row = "deepskyblue"), html_headers = NULL,
- html_change_col_name = "chng_type", html_group_col_name = "grp",
round_output_to = 3)
}
\arguments{
@@ -22,8 +19,6 @@ by which to group_by.}
\item{exclude}{The columns which should be excluded from the comparison}
-\item{limit_html}{maximum number of rows to show in the html diff. >1000 not recommended}
-
\item{tolerance}{The amount in fraction to which changes are ignored while showing the
visual representation. By default, the value is 0 and any change in the value of variables
is shown off. Doesn't apply to categorical variables.}
@@ -36,15 +31,6 @@ is shown off. Doesn't apply to categorical variables.}
\item{keep_unchanged_cols}{whether to preserve unchanged values or not. Defaults to \code{TRUE}}
-\item{color_scheme}{What color scheme to use for the HTML output. Should be a vector/list with
-named_elements. Default - \code{c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")}}
-
-\item{html_headers}{A character vector of column names to be used in the table. Defaults to \code{colnames}.}
-
-\item{html_change_col_name}{Name of the change column to use in the HTML table. Defaults to \code{chng_type}.}
-
-\item{html_group_col_name}{Name of the group column to be used in the table (if there are multiple grouping vars). Defaults to \code{grp}.}
-
\item{round_output_to}{Number of digits to round the output to. Defaults to 3.}
}
\description{
diff --git a/man/create_output_table.Rd b/man/create_output_table.Rd
new file mode 100644
index 0000000..b4229ce
--- /dev/null
+++ b/man/create_output_table.Rd
@@ -0,0 +1,33 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/fnsOutputs.R
+\name{create_output_table}
+\alias{create_output_table}
+\title{Create human readable output from the comparison_df output}
+\usage{
+create_output_table(comparison_output, output_type = "html",
+ file_name = NULL, limit = 100, color_scheme = c(addition =
+ "#52854C", removal = "#FC4E07", unchanged_cell = "#999999", unchanged_row
+ = "#293352"), headers = NULL, change_col_name = "chng_type",
+ group_col_name = "grp")
+}
+\arguments{
+\item{comparison_output}{Output from the comparison Table functions}
+
+\item{output_type}{Type of comparison output. Defaults to `html`}
+
+\item{file_name}{Where to write the output to. Default to NULL which output to the Rstudio viewer (not supported for `xlsx`)}
+
+\item{limit}{maximum number of rows to show in the diff. >1000 not recommended for HTML}
+
+\item{color_scheme}{What color scheme to use for the output. Should be a vector/list with
+named_elements. Default - \code{c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")}}
+
+\item{headers}{A character vector of column names to be used in the table. Defaults to \code{colnames}.}
+
+\item{change_col_name}{Name of the change column to use in the table. Defaults to \code{chng_type}.}
+
+\item{group_col_name}{Name of the group column to be used in the table (if there are multiple grouping vars). Defaults to \code{grp}.}
+}
+\description{
+Currently `html` and `xlsx` are supported
+}
diff --git a/man/view_html.Rd b/man/view_html.Rd
index 43724b6..17313e1 100644
--- a/man/view_html.Rd
+++ b/man/view_html.Rd
@@ -1,5 +1,5 @@
% Generated by roxygen2: do not edit by hand
-% Please edit documentation in R/fnsComparison.R
+% Please edit documentation in R/fnsOutputs.R
\name{view_html}
\alias{view_html}
\title{View Comparison output HTML}
diff --git a/tests/testthat/test-fnsComparison.R b/tests/testthat/test-fnsComparison.R
index e2f1ad4..f8193fd 100644
--- a/tests/testthat/test-fnsComparison.R
+++ b/tests/testthat/test-fnsComparison.R
@@ -150,16 +150,9 @@ expected_comparison_df = data.frame(grp = c(3, 4),
# arrange(desc(chng_type)) %>% arrange_("var1")
expect_equal(ctable$comparison_df, expected_comparison_df)
-#===============================================================================
-# Limit
-context("compare_df: limit")
-max_rows = 2
-ctable = compare_df(new_df, old_df, c("var1", "var2"), limit_html = max_rows)
-expect_equal(ctable$html_output %>% as.character() %>% stringr::str_count("% str_replace_all("\\n", "")
-
- html_output_string %>%
- str_extract("thead.*thead") %>%
- str_extract_all("'>.+?<") %>%
- magrittr::extract2(1) %>%
- str_replace_all("'>(.*)<", "\\1")
-
-}
-
-context("compare_df: Headers")
-
-test_that("compare_df: headers with 1 grouping column", {
- ctable = compare_df(new_df, old_df, c("var1"),
- html_headers = c(var1 = "Variable 1", var2 = "Variable 2", val1 = "Value 1", val2 = "Value 2", val3 = "Value 3"))
-
- expected_headers = c("Variable 1", "chng_type", "Variable 2",
- "Value 1", "Value 2", "Value 3")
- expect_equal(expected_headers, get_html_header_names(ctable))
-})
-
-
-
-test_that("compare_df: headers with partial matching", {
- ctable = compare_df(new_df, old_df, c("var1"),
- html_headers = c(var1 = "Variable 1", val1 = "Value 1", val3 = "Value 3"))
-
- expected_headers = c("Variable 1", "chng_type", "var2",
- "Value 1", "val2", "Value 3")
- expect_equal(expected_headers, get_html_header_names(ctable))
-})
-
-test_that("compare_df: headers with additional matching", {
- ctable = compare_df(new_df, old_df, c("var1"),
- html_headers = c(var1 = "Variable 1", var2 = "Variable 2", val1 = "Value 1", val2 = "Value 2", val4 = "Value 4"))
-
- expected_headers = c("Variable 1", "chng_type", "Variable 2",
- "Value 1", "Value 2", "val3")
- expect_equal(expected_headers, get_html_header_names(ctable))
-
-})
-
-test_that("compare_df: headers and group column and change column", {
- ctable = compare_df(new_df, old_df, c("var1"),
- html_headers = c(var1 = "Variable 1", var2 = "Variable 2", val1 = "Value 1", val2 = "Value 2", val3 = "Value 3"),
- html_group_col_name = "Group ID", html_change_col_name = "Type of Change")
- expected_headers = c("Variable 1", "Type of Change", "Variable 2",
- "Value 1", "Value 2", "Value 3")
- expect_equal(expected_headers, get_html_header_names(ctable))
-})
-
-test_that("compare_df: only group column and change column", {
- ctable = compare_df(new_df, old_df, c("var1"),
- html_group_col_name = "Group ID", html_change_col_name = "Type of Change")
-
- expected_headers = c("var1", "Type of Change", "var2",
- "val1", "val2", "val3")
- expect_equal(expected_headers, get_html_header_names(ctable))
-
-
-})
-
-test_that("compare_df: headers with more than 1 grouping column and group column and change column", {
-
- ctable = compare_df(new_df, old_df, c("var1", "var2"),
- html_headers = c(var1 = "Variable 1", var2 = "Variable 2", val1 = "Value 1", val2 = "Value 2", val3 = "Value 3"),
- html_change_col_name = "Type of Change", html_group_col_name = "Group ID")
-
- expected_headers = c("Group ID", "Type of Change", "Variable 1", "Variable 2",
- "Value 1", "Value 2", "Value 3")
-
- expect_equal(expected_headers, get_html_header_names(ctable))
-})
-
#===============================================================================
context("compare_df: Integration Edge case")
diff --git a/tests/testthat/test-fnsOutputs.R b/tests/testthat/test-fnsOutputs.R
new file mode 100644
index 0000000..4180f80
--- /dev/null
+++ b/tests/testthat/test-fnsOutputs.R
@@ -0,0 +1,180 @@
+
+library(testthat)
+library(dplyr)
+library(compareDF)
+library(stringr)
+
+old_df = data.frame(var1 = c("A", "B", "C"),
+ var2 = c("Z", "Y", "X"),
+ val1 = c(1, 2, 3),
+ val2 = c("A1", "B1", "C1"),
+ val3 = c(1, 2, 3)
+)
+
+new_df = data.frame(var1 = c("A", "B", "C"),
+ var2 = c("Z", "Y", "X"),
+ val1 = c(1, 2, 3),
+ val2 = c("A1", "B1", "C2"),
+ val3 = c(1, 2.1, 4)
+)
+
+#===============================================================================
+# HTML
+#===============================================================================
+# Limit
+context("compare_df: limit")
+max_rows = 2
+ctable = compare_df(new_df, old_df, c("var1", "var2"))
+html_output = create_output_table(ctable, limit = max_rows)
+expect_equal(html_output %>% as.character() %>% stringr::str_count("
%
+ str_extract("thead.*thead") %>%
+ str_extract_all("'>.+?<") %>%
+ magrittr::extract2(1) %>%
+ str_replace_all("'>(.*)<", "\\1")
+
+}
+
+context("compare_df: Headers")
+
+test_that("compare_df: headers with 1 grouping column", {
+ ctable = compare_df(new_df, old_df, c("var1"))
+ html_headers = c(var1 = "Variable 1", var2 = "Variable 2", val1 = "Value 1", val2 = "Value 2", val3 = "Value 3")
+ html_output = create_output_table(ctable, headers = html_headers)
+
+ expected_headers = c("Variable 1", "chng_type", "Variable 2",
+ "Value 1", "Value 2", "Value 3")
+ expect_equal(expected_headers, get_html_header_names(html_output))
+})
+
+
+
+test_that("compare_df: headers with partial matching", {
+ ctable = compare_df(new_df, old_df, c("var1"))
+ html_headers = c(var1 = "Variable 1", val1 = "Value 1", val3 = "Value 3")
+ html_output = create_output_table(ctable, headers = html_headers)
+
+ expected_headers = c("Variable 1", "chng_type", "var2",
+ "Value 1", "val2", "Value 3")
+ expect_equal(expected_headers, get_html_header_names(html_output))
+})
+
+test_that("compare_df: headers with additional matching", {
+ ctable = compare_df(new_df, old_df, c("var1"))
+ html_headers = c(var1 = "Variable 1", var2 = "Variable 2", val1 = "Value 1", val2 = "Value 2", val4 = "Value 4")
+ html_output = create_output_table(ctable, headers = html_headers)
+
+ expected_headers = c("Variable 1", "chng_type", "Variable 2",
+ "Value 1", "Value 2", "val3")
+ expect_equal(expected_headers, get_html_header_names(html_output))
+
+})
+
+test_that("compare_df: headers and group column and change column", {
+ ctable = compare_df(new_df, old_df, c("var1"))
+ html_headers = c(var1 = "Variable 1", var2 = "Variable 2", val1 = "Value 1", val2 = "Value 2", val3 = "Value 3")
+ html_group_col_name = "Group ID"
+ html_change_col_name = "Type of Change"
+ html_output = create_output_table(ctable, headers = html_headers, group_col_name = html_group_col_name, change_col_name = html_change_col_name)
+
+ expected_headers = c("Variable 1", "Type of Change", "Variable 2",
+ "Value 1", "Value 2", "Value 3")
+ expect_equal(expected_headers, get_html_header_names(html_output))
+})
+
+test_that("compare_df: only group column and change column", {
+ ctable = compare_df(new_df, old_df, c("var1"))
+ html_group_col_name = "Group ID"
+ html_change_col_name = "Type of Change"
+ html_output = create_output_table(ctable, group_col_name = html_group_col_name, change_col_name = html_change_col_name)
+
+ expected_headers = c("var1", "Type of Change", "var2",
+ "val1", "val2", "val3")
+ expect_equal(expected_headers, get_html_header_names(html_output))
+
+
+})
+
+test_that("compare_df: headers with more than 1 grouping column and group column and change column", {
+
+ ctable = compare_df(new_df, old_df, c("var1", "var2"))
+ html_group_col_name = "Group ID"
+ html_change_col_name = "Type of Change"
+ html_headers = c(var1 = "Variable 1", var2 = "Variable 2", val1 = "Value 1", val2 = "Value 2", val3 = "Value 3")
+ html_output = create_output_table(ctable, headers = html_headers, group_col_name = html_group_col_name, change_col_name = html_change_col_name)
+
+ expected_headers = c("Group ID", "Type of Change", "Variable 1", "Variable 2",
+ "Value 1", "Value 2", "Value 3")
+
+ expect_equal(expected_headers, get_html_header_names(html_output))
+})
+
+test_that("compare_df: write output to file", {
+
+ ctable = compare_df(new_df, old_df, c("var1", "var2"))
+ temp_file = tempfile()
+ html_output = create_output_table(ctable, file_name = temp_file)
+ expect_true(file.exists(temp_file))
+ unlink(temp_file)
+})
+
+
+#===============================================================================
+# XLSX
+#===============================================================================
+
+context("compare_df: Output to Excel")
+
+old_df = data.frame(var1 = c("A", "B", "C"),
+ var2 = c("Z", "Y", "X"),
+ val1 = c(1, 2, 3),
+ val2 = c("A1", "B1", "C1"),
+ val3 = c(1, 2, 3)
+)
+
+new_df = data.frame(var1 = c("A", "B", "C"),
+ var2 = c("Z", "Y", "X"),
+ val1 = c(1, 2, 3),
+ val2 = c("A1", "B1", "C2"),
+ val3 = c(1, 2.1, 4)
+)
+
+compare_output = compareDF::compare_df(old_df, new_df, c('var1', 'var2'))
+
+test_that("compare_df: Error out if file name is NULL", {
+ expect_error(create_output_table(compare_output, output_type = 'xlsx'), "file_name cannot be null if output format is xlsx")
+})
+
+
+test_that("compare_df: Write to file correctly", {
+ temp_file = tempfile()
+ create_output_table(compare_output, output_type = 'xlsx', file_name = temp_file)
+ expect_true(file.exists(temp_file))
+ unlink(temp_file)
+})
+
+# context("compare_df: Test Large output")
+#
+# old_df = data.frame(var1 = paste(1:12000, c("A"), sep = "_"),
+# var2 = c("Z", "Y", "X"),
+# val1 = c(1, 2, 3),
+# val2 = c("A1", "B1", "C1"),
+# val3 = c(1, 2, 3)
+# )
+#
+# new_df = data.frame(var1 = paste(1:9000, c("A"), sep = "_"),
+# var2 = c("Z", "Y", "X"),
+# val1 = c(1, 5, 3),
+# val2 = c("A1", "B1", "C2"),
+# val3 = c(1, 2, 3)
+# )
+# big_output = compareDF::compare_df(old_df, new_df, c('var1', 'var2'))
+# create_output_table(big_output, output_type = 'xlsx', file_name = "test_file.xlsx")
+
 @@ -109,11 +109,16 @@ scores, which are in _Discipline_, _Maths_, and _Maths_ respectively.
_Dhakkan_ and _Mugger_ have dropped out of the dataset from 2010 and the all the columns for the rows are shown
in red, which _DikChik_ and _Vikram_ have joined new in the data set and all the columns for the rows are in green.
-The same data is represented in tabular form (for further analysis, if necessary) in the
-`comparison_table_diff` object
+```{r}
+create_output_table(ctable_student)
+```
+
+### XLSX Output
+
+Alternately you can write to an xlsx file as well
```{r}
-ctable_student$comparison_table_diff
+create_output_table(ctable_student, output_type = 'xlsx', file_name = "test_file.xlsx")
```
## Change Count and Summary
@@ -129,7 +134,7 @@ ctable_student$change_count
ctable_student$change_summary
```
-## Grouping Multiple Columns
+### Grouping Multiple Columns
We can also group_multiple columns into the grouping variable
@@ -142,18 +147,17 @@ ctable_student_div$comparison_df
Now _Rohits_ in each individual division are considered as belonging to separate
groups and compared accordingly. All the other summaries also change appropriately.
-## Excluding certain Columns
+### Excluding certain Columns
-You can ignore certain columns using the *exclude* parameter. The fields that have to be
+You can ignore certain columns using the _exclude_ parameter. The fields that have to be
excluded can be given as a character vector. (This is a convenience function to deal with
the case where some columns are not included)
-## Preserving all rows
+### Preserving all rows
The default behavior of the `compare_df` function is to show only the records that have changed. Sometimes the use might want to preserve all the records even after the comparison and not just see the values that have been changed (especially in the HTML). In this case, you can set the `keep_unchanged` parameter to `TRUE`.
-
-## Limiting HTML size
+### Limiting HTML size
For dataframes which have a large amount of differences in them, generating HTML might take
a long time and crash your system. So the maximum diff size for the HTML (and for the HTML
@@ -161,12 +165,11 @@ visualization only) is capped at 100 by default. If you want to see more differe
the `limit_html` parameter appropriately. NOTE: This is only of the HTML output which is used for visual
checking. The main comparison data frame and the summaries ALWAYS include data from all the rows.
-## Changing HTML color
-
-You can use the `color_scheme` parameter to change the color of the HTML generated. The parameter must me a named vector or a list with the appropriate names - The default values are `c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")` but can be changed as needed by the user.
+### Changing color
+You can use the `color_scheme` parameter to change the color of the cells generated. The parameter must me a named vector or a list with the appropriate names - The default values are `c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")` but can be changed as needed by the user.
-## Tolerance
+### Tolerance
It is possible that you'd like numbers very close to each other to be ignored. For example,
if the marks of a student vary by less that 5% across the years, it could be due to random
@@ -176,7 +179,7 @@ less than 5% apart from the lower value.
```{r, results = 'hide'}
ctable_student_div = compare_df(results_2011, results_2010, c("Division", "Student"), tolerance = 0.05)
-ctable_student_div$html_output
+create_output_table(ctable_student_div)
```
@@ -109,11 +109,16 @@ scores, which are in _Discipline_, _Maths_, and _Maths_ respectively.
_Dhakkan_ and _Mugger_ have dropped out of the dataset from 2010 and the all the columns for the rows are shown
in red, which _DikChik_ and _Vikram_ have joined new in the data set and all the columns for the rows are in green.
-The same data is represented in tabular form (for further analysis, if necessary) in the
-`comparison_table_diff` object
+```{r}
+create_output_table(ctable_student)
+```
+
+### XLSX Output
+
+Alternately you can write to an xlsx file as well
```{r}
-ctable_student$comparison_table_diff
+create_output_table(ctable_student, output_type = 'xlsx', file_name = "test_file.xlsx")
```
## Change Count and Summary
@@ -129,7 +134,7 @@ ctable_student$change_count
ctable_student$change_summary
```
-## Grouping Multiple Columns
+### Grouping Multiple Columns
We can also group_multiple columns into the grouping variable
@@ -142,18 +147,17 @@ ctable_student_div$comparison_df
Now _Rohits_ in each individual division are considered as belonging to separate
groups and compared accordingly. All the other summaries also change appropriately.
-## Excluding certain Columns
+### Excluding certain Columns
-You can ignore certain columns using the *exclude* parameter. The fields that have to be
+You can ignore certain columns using the _exclude_ parameter. The fields that have to be
excluded can be given as a character vector. (This is a convenience function to deal with
the case where some columns are not included)
-## Preserving all rows
+### Preserving all rows
The default behavior of the `compare_df` function is to show only the records that have changed. Sometimes the use might want to preserve all the records even after the comparison and not just see the values that have been changed (especially in the HTML). In this case, you can set the `keep_unchanged` parameter to `TRUE`.
-
-## Limiting HTML size
+### Limiting HTML size
For dataframes which have a large amount of differences in them, generating HTML might take
a long time and crash your system. So the maximum diff size for the HTML (and for the HTML
@@ -161,12 +165,11 @@ visualization only) is capped at 100 by default. If you want to see more differe
the `limit_html` parameter appropriately. NOTE: This is only of the HTML output which is used for visual
checking. The main comparison data frame and the summaries ALWAYS include data from all the rows.
-## Changing HTML color
-
-You can use the `color_scheme` parameter to change the color of the HTML generated. The parameter must me a named vector or a list with the appropriate names - The default values are `c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")` but can be changed as needed by the user.
+### Changing color
+You can use the `color_scheme` parameter to change the color of the cells generated. The parameter must me a named vector or a list with the appropriate names - The default values are `c("addition" = "green", "removal" = "red", "unchanged_cell" = "gray", "unchanged_row" = "deepskyblue")` but can be changed as needed by the user.
-## Tolerance
+### Tolerance
It is possible that you'd like numbers very close to each other to be ignored. For example,
if the marks of a student vary by less that 5% across the years, it could be due to random
@@ -176,7 +179,7 @@ less than 5% apart from the lower value.
```{r, results = 'hide'}
ctable_student_div = compare_df(results_2011, results_2010, c("Division", "Student"), tolerance = 0.05)
-ctable_student_div$html_output
+create_output_table(ctable_student_div)
```
 @@ -186,13 +189,13 @@ diff calculation or in the output
Naturally, tolerance has no meaning for non-numeric values.
## Additional features
-- set the color scheme in the HTML using the `color_scheme` argument
-- preserve the rows that have not changed in the anlysis using the `keep_unchanged_rows` argument
-- use `difference` as an argument to use difference rather than ratio for tolerance
-- options to name the columns in the HTML output
-- option change column name
-- option to change group column name
-- keep only the columns which have changed using `keep_unchanged_cols`
+* set the color scheme in the HTML using the `color_scheme` argument
+* preserve the rows that have not changed in the analysis using the `keep_unchanged_rows` argument
+* use `difference` as an argument to use difference rather than ratio for tolerance
+* options to name the columns in the HTML output
+* option change column name
+* option to change group column name
+* keep only the columns which have changed using `keep_unchanged_cols`
## Using compare DF in GAP analysis
@@ -203,8 +206,8 @@ The compareDF package can be used to conduct effective Gap analyses. If the pack
Thanks to Nitin for proofreading the doc and making sure everything made sense.
## Contributors
-- Brice Richard
-- Joshua David Barillas - https://github.com/jdbarillas
+* Brice Richard
+* Joshua David Barillas -
@@ -186,13 +189,13 @@ diff calculation or in the output
Naturally, tolerance has no meaning for non-numeric values.
## Additional features
-- set the color scheme in the HTML using the `color_scheme` argument
-- preserve the rows that have not changed in the anlysis using the `keep_unchanged_rows` argument
-- use `difference` as an argument to use difference rather than ratio for tolerance
-- options to name the columns in the HTML output
-- option change column name
-- option to change group column name
-- keep only the columns which have changed using `keep_unchanged_cols`
+* set the color scheme in the HTML using the `color_scheme` argument
+* preserve the rows that have not changed in the analysis using the `keep_unchanged_rows` argument
+* use `difference` as an argument to use difference rather than ratio for tolerance
+* options to name the columns in the HTML output
+* option change column name
+* option to change group column name
+* keep only the columns which have changed using `keep_unchanged_cols`
## Using compare DF in GAP analysis
@@ -203,8 +206,8 @@ The compareDF package can be used to conduct effective Gap analyses. If the pack
Thanks to Nitin for proofreading the doc and making sure everything made sense.
## Contributors
-- Brice Richard
-- Joshua David Barillas - https://github.com/jdbarillas
+* Brice Richard
+* Joshua David Barillas -