The external validation experiment of aki phenogroup research
We assume you are already able to access the MIMIC-III (V1.4), which is a publicily available electronic health record (EHR) databse which contains over 40 thousand patients who stayed in critical care units of the Beth Israel Deaconess Medical Center between 2001 and 2012.
If you are not able to access MIMIC-III, please complete a required training course and then submit an application at request link. Once the application is approved, you can download the MIMIC-III freely.
Please ensure your computer has enough storage space because MIMIC-III is a large dataset (45 GiB is required in this study)
Please install Python 3.7 environment, as well as lifelines (Version 0.24.8), matplotlib (3.2.1), numpy (1.18.4), pandas (1.0.4), scikit-learn (0.23.1), joblib (0.15.1), and scipy (1.4.1) packages in advance.
Please clone the project, and then uncompress the entire MIMIC-III dataset into:
/resource/raw_data/
The files in the folder are like:
/resource/raw_data/...
ADMISSIONS.csv...
CALLOUT.csv...
......
TRANSFERS.csv
Please run the 'read_raw_mimic_data.py' script in the /src folder.
This script is responsible to reconstruct the dataset to a structured format and extract AKI and in-hospital mortality events. As the MIMIC-III is a large dataset, this script typically needs several hours to parse the entire dataset. Once the script is executed successfully, we can find a file named 'mimic_unpreprocessed.csv' in the /resource folder. The file contains 46,521 admissions.
Please run the 'visit_and_feature_filter.py' script in the /src folder
According to the requirement of this study, we need to included adult heart failure patients with normal renal function and relative complete EHR data to conduct analysis. Therefore, discarding undesirable information contained in the 'mimic_unpreprocessed.csv' is necessary.
The 'visit_and_feature_filter.py' script is responsible to discard undesirable information, which basically follows the procedure in Figure S2 in the paper. Once the script is successfully executed, we can find a file named 'filtered.csv' in the /resource folder. The file contains 1,006 admissions, and each admission consists of 39 variables.
Please run the 'value_convert.py' script This script is responsible to complete two tasks.
- Converting all continuous variables to new values which follow standard normal distribution
- If the value of a variable is missing, imputing the mean of the two phenogroups, as described in the paper.
Once the script is successfully executed, we can find a file named 'preprocessed_data.csv' in the /resource folder. All missing values in the 'filtered.csv' are imputed in this file.
Please run the 'phenogroup_assignment.py' script.
As described in the paper, the phenogroup assignment of a patient is decided by its distance to the centroids of two phenogroups.
Once the script is successfully executed, we can find a file named 'phenogroup_assignment.csv' in the /resource folder.
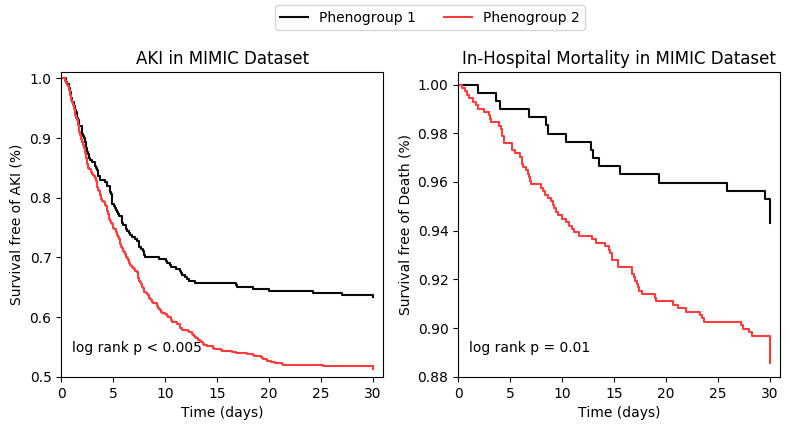
Please run the 'kaplan_meier_curve.py' script.
The script is responsible to plot the survival curve of two phenogroups assigned in last step.
Once the script is successfully executed, we can find a file named 'kaplan_meier_curve.png' in the /resource folder. The figure is same as the Figure 2 in our paper.
Please run the 'value_recover_with_phenogroup.py' script. This script is responsible to recover the value of variables. Note the missing value will also be recovered to the mean of centroids. Once the script is successfully executed, we can find a file named 'recovered_data_with_group.csv' in the /resource folder. The medians, frequencies, as well as the quartiles and counts of MIMIC dataset listed in Table S2 were summarized by this file.
Please run the 'transfer_test.py' script This script is responsible test the robustness of our risk stratification model. Once the script is successfully executed, we can see the output like:
aki_only_group, recall (sensitivity): 0.75991, specificity: 0.34239, auc: 0.55115
aki_10, recall (sensitivity): 0.37445, specificity: 0.65217, auc: 0.53239
aki_10_group, recall (sensitivity): 0.47797, specificity: 0.56159, auc: 0.54564
aki_39, recall (sensitivity): 0.54405, specificity: 0.55978, auc: 0.57027
aki_39_group, recall (sensitivity): 0.57269, specificity: 0.53986, auc: 0.57440
death_only_group, recall (sensitivity): 0.82653, specificity: 0.30947, auc: 0.56800
death_10, recall (sensitivity): 0.53061, specificity: 0.67181, auc: 0.62221
death_10_group, recall (sensitivity): 0.62245, specificity: 0.59912, auc: 0.64722
death_39, recall (sensitivity): 0.48980, specificity: 0.74449, auc: 0.64522
death_39_group, recall (sensitivity): 0.52041, specificity: 0.72026, auc: 0.64457
The results is same as the MIMIC dataset results that we listed in the Table 3.