diff --git a/.travis.yml b/.travis.yml
index 19518d7..8a2f174 100644
--- a/.travis.yml
+++ b/.travis.yml
@@ -19,7 +19,7 @@ matrix:
before_script: Rscript -e "remotes::install_github('ludvigolsen/xpectr')"
- r: release
name: multiple-devel
- before_script: Rscript -e "remotes::install_github(c('ludvigolsen/xpectr', 'r-lib/vctrs', 'tidyverse/dplyr', 'tidyverse/tidyr', 'tidyverse/tibble'), dependencies = TRUE)"
+ before_script: Rscript -e "remotes::install_github(c('ludvigolsen/xpectr', 'tidyverse/dplyr', 'tidyverse/tidyr', 'tidyverse/tibble'), dependencies = TRUE)"
- r: release
os: osx
name: release osx
diff --git a/CRAN-RELEASE b/CRAN-RELEASE
deleted file mode 100644
index b24f3f3..0000000
--- a/CRAN-RELEASE
+++ /dev/null
@@ -1,2 +0,0 @@
-This package was submitted to CRAN on 2020-10-17.
-Once it is accepted, delete this file and tag the release (commit 03ceacf).
diff --git a/DESCRIPTION b/DESCRIPTION
index 6f6e2d8..7457b1e 100644
--- a/DESCRIPTION
+++ b/DESCRIPTION
@@ -1,6 +1,6 @@
Package: rearrr
Title: Rearranging Data
-Version: 0.1.0
+Version: 0.1.0.9000
Authors@R:
c(person(given = "Ludvig Renbo",
family = "Olsen",
@@ -23,6 +23,7 @@ Imports:
purrr (>= 0.3.4),
rlang (>= 0.4.7),
stats,
+ tibble,
utils
Suggests:
covr,
diff --git a/NAMESPACE b/NAMESPACE
index a1395e7..190f229 100644
--- a/NAMESPACE
+++ b/NAMESPACE
@@ -44,6 +44,7 @@ export(rotate_2d)
export(rotate_3d)
export(shear_2d)
export(shear_3d)
+export(shuffle_hierarchy)
export(square)
export(swirl_2d)
export(swirl_3d)
diff --git a/NEWS.md b/NEWS.md
index 237c7c3..be35242 100644
--- a/NEWS.md
+++ b/NEWS.md
@@ -1,4 +1,10 @@
+# rearrr 0.2.0
+
+* Adds `shuffle_hierarchy()` for shuffling a multi-column hierarchy of groups. (Rearranger)
+
+* Adds optional recursion to `pair_extremes()`. This adds the `num_pairings` and `balance` arguments. Now returns one additional sorting factor per `num_pairing` level.
+
# rearrr 0.1.0
Note: Multiple of the new functions also have `*_vec()` versions that take and return vectors. The same can *usually* be achieved with the regular versions, but these wrappers make it easier and less verbose.
diff --git a/R/pair_extremes.R b/R/pair_extremes.R
index 00fdd2d..ded904d 100644
--- a/R/pair_extremes.R
+++ b/R/pair_extremes.R
@@ -76,6 +76,9 @@
#' # Shuffle the order of the pairs
#' pair_extremes(df, col = "A", shuffle_pairs = TRUE)
#'
+#' # Use recursive pairing
+#' pair_extremes(df, col = "A", num_pairings = 2)
+#'
#' # Grouped by G
#' df %>%
#' dplyr::select(G, A) %>% # For clarity
@@ -100,16 +103,18 @@
pair_extremes <- function(data,
col = NULL,
unequal_method = "middle",
- # num_pairings = 1, # TODO
+ num_pairings = 1,
+ balance = "mean",
shuffle_members = FALSE,
shuffle_pairs = FALSE,
- factor_name = ".pair",
+ factor_name = ifelse(num_pairings == 1, ".pair", ".pairing"),
overwrite = FALSE) {
extreme_pairing_rearranger_(
data = data,
col = col,
unequal_method = unequal_method,
- num_pairings = 1,
+ num_pairings = num_pairings,
+ balance = balance,
shuffle_members = shuffle_members,
shuffle_pairs = shuffle_pairs,
factor_name = factor_name,
@@ -121,6 +126,8 @@ pair_extremes <- function(data,
#' @export
pair_extremes_vec <- function(data,
unequal_method = "middle",
+ num_pairings = 1,
+ balance = "mean",
shuffle_members = FALSE,
shuffle_pairs = FALSE){
checkmate::assert(checkmate::check_vector(data, strict = TRUE),
@@ -128,6 +135,8 @@ pair_extremes_vec <- function(data,
pair_extremes(
data = data,
unequal_method = unequal_method,
+ num_pairings = num_pairings,
+ balance = balance,
shuffle_members = shuffle_members,
shuffle_pairs = shuffle_pairs,
factor_name = NULL,
diff --git a/R/rearrange_factors.R b/R/rearrange_factors.R
index 115241e..cf79ac4 100644
--- a/R/rearrange_factors.R
+++ b/R/rearrange_factors.R
@@ -29,19 +29,25 @@ create_rearrange_factor_pair_extremes_ <- function(size, unequal_method = "middl
if (size == 1) {
return(1)
}
+
half_size <- floor(size / 2)
idx <- seq_len(half_size)
+
if (half_size * 2 == size) {
return(c(idx, rev(idx)))
- } else {
- if (unequal_method == "middle") {
- middle <- ceiling((half_size / 2)) + 1
- idx <- ifelse(idx >= middle, idx + 1, idx)
- return(c(idx, middle, rev(idx)))
- } else if (unequal_method == "first") {
- return(c(1, c(idx, rev(idx)) + 1))
- } else if (unequal_method == "last") {
- return(c(c(idx, rev(idx)), max(idx) + 1))
- }
+ }
+
+ if (unequal_method == "middle") {
+ middle <- ceiling(half_size / 2) + 1
+ idx <- ifelse(idx >= middle, idx + 1, idx)
+ return(c(idx, middle, rev(idx)))
+ }
+
+ if (unequal_method == "first") {
+ return(c(1, c(idx, rev(idx)) + 1))
+ }
+
+ if (unequal_method == "last") {
+ return(c(c(idx, rev(idx)), max(idx) + 1))
}
}
diff --git a/R/rearrange_methods.R b/R/rearrange_methods.R
index 92809fb..0ed97e0 100644
--- a/R/rearrange_methods.R
+++ b/R/rearrange_methods.R
@@ -320,10 +320,12 @@ rearrange_by_distance <- function(data,
# TODO Add aggregate_fn for recursive pairings
-rearrange_pair_extremes <- function(data, cols,
+rearrange_pair_extremes <- function(data,
+ cols,
overwrite,
unequal_method,
num_pairings,
+ balance,
shuffle_members,
shuffle_pairs,
factor_name,
@@ -335,43 +337,165 @@ rearrange_pair_extremes <- function(data, cols,
return(data)
}
- # Order data frame
- data <- data[order(data[[col]]), , drop = FALSE]
+ equal_nrows <- nrow(data) %% 2 == 0
+
+ # Prepare factor names for output
+ factor_names <- factor_name
+ if (!is.null(factor_names) && num_pairings > 1){
+ factor_names <- paste0(factor_name, "_", seq_len(num_pairings))
+ }
## First extreme pairing
- # Create
- local_tmp_rearrange_var <- create_tmp_var(data, ".tmp_rearrange_col")
- data[[local_tmp_rearrange_var]] <-
- create_rearrange_factor_pair_extremes_(
- size = nrow(data), unequal_method = unequal_method
- )
+ # Create num_pairings tmp column names
+ tmp_rearrange_vars <- purrr::map(.x = seq_len(num_pairings), .f = ~ {
+ create_tmp_var(data, paste0(".tmp_rearrange_col_", .x))
+ }) %>% unlist(recursive = TRUE)
+
+ # Pair the extreme values and order by the pairs
+ data <- order_and_pair_extremes_(
+ data = data,
+ col = col,
+ new_col = tmp_rearrange_vars[[1]],
+ unequal_method = unequal_method
+ )
# Order data by the pairs
data <- order_by_group(
data = data,
- group_col = local_tmp_rearrange_var,
- shuffle_members = shuffle_members,
- shuffle_pairs = shuffle_pairs
+ group_col = tmp_rearrange_vars[[1]],
+ shuffle_members = FALSE, # Done at the end
+ shuffle_pairs = FALSE # Done at the end
)
# TODO Perform recursive pairing
- # if (num_pairings > 1){
- #
- #
- #
- # }
+ if (num_pairings > 1){
+
+ # Get environment so we can update `data`
+ pairing_env <- environment()
+
+ # Perform num_pairings-1 pairings
+ plyr::l_ply(seq_len(num_pairings - 1), function(i) {
+
+ # Note that `tmp_rearrange_vars[[i]]` is for the previous level
+ # and `tmp_rearrange_vars[[i + 1]]` is for the current level
+
+ # What to balance ("mean", "spread", "min", or "max")
+ if (length(balance) > 1) {
+ current_balance_target <- balance[[i]]
+ } else {
+ current_balance_target <- balance
+ }
+
+ # Define function to summarize with
+ if (current_balance_target == "mean") {
+ summ_fn <- sum
+ } else if (current_balance_target == "spread") {
+ summ_fn <- function(v) {
+ sum(abs(diff(v)))
+ }
+ } else if (current_balance_target == "min") {
+ summ_fn <- min
+ } else if (current_balance_target == "max") {
+ summ_fn <- max
+ }
+
+ # Aggregate values for pairs from previous pairing
+ tmp_group_scores <- data %>%
+ dplyr::group_by(!!as.name(tmp_rearrange_vars[[i]])) %>%
+ dplyr::summarize(group_aggr = summ_fn(!!as.name(col)))
+
+ if (!equal_nrows & unequal_method == "first") {
+
+ # Reorder with first group always first
+ # (otherwise doesn't work with negative numbers)
+ tmp_group_scores_sorted <- tmp_group_scores %>%
+ dplyr::filter(dplyr::row_number() == 1) %>%
+ dplyr::bind_rows(
+ tmp_group_scores %>%
+ dplyr::filter(dplyr::row_number() != 1) %>%
+ dplyr::arrange(.data$group_aggr))
+ } else {
+ tmp_group_scores_sorted <- tmp_group_scores %>%
+ dplyr::arrange(.data$group_aggr)
+ }
+
+ # Pair the extreme pairs and order by the new pairs
+ tmp_rearrange <- order_and_pair_extremes_(
+ data = tmp_group_scores_sorted,
+ col = "group_aggr",
+ new_col = tmp_rearrange_vars[[i + 1]],
+ unequal_method = unequal_method
+ ) %>%
+ base_select_(cols = c(tmp_rearrange_vars[[i]], tmp_rearrange_vars[[i + 1]]))
+
+ # Add the new pair identifiers to `data`
+ data <- data %>%
+ dplyr::left_join(tmp_rearrange, by = tmp_rearrange_vars[[i]])
+
+ # Order data by the pairs
+ data <- order_by_group(
+ data = data,
+ group_col = tmp_rearrange_vars[[i + 1]],
+ shuffle_members = FALSE,
+ shuffle_pairs = FALSE
+ )
- # TODO Make this work for num_pairings factors
+ # Update `data` in parent environment
+ pairing_env[["data"]] <- data
- # Add rearrange factor
- data <- add_info_col_(
- data = base_deselect_(data, cols = local_tmp_rearrange_var),
- nm = factor_name,
- content = as.factor(data[[local_tmp_rearrange_var]]),
- overwrite = overwrite
- )
+ })
+ }
+
+ # Find columns to shuffle
+ shuffling_group_cols <- rev(tmp_rearrange_vars)
+ cols_to_shuffle <- c()
+ if (isTRUE(shuffle_members))
+ cols_to_shuffle <- shuffling_group_cols
+ if (isTRUE(shuffle_pairs)) {
+ shuffling_group_cols <- c(shuffling_group_cols, col)
+ cols_to_shuffle <- c(cols_to_shuffle, col)
+ }
+
+ # Shuffle members and/or pairs if specified
+ if (length(cols_to_shuffle) > 0) {
+ data <- dplyr::ungroup(data) %>%
+ shuffle_hierarchy(group_cols = shuffling_group_cols,
+ cols_to_shuffle = cols_to_shuffle,
+ leaf_has_groups = !shuffle_members)
+ }
+
+ if (!is.null(factor_names)){
+ # Convert to factors and give correct names
+ rearrange_factors <- as.list(data[, tmp_rearrange_vars, drop=FALSE])
+ rearrange_factors <- purrr::map(rearrange_factors, .f = ~{as.factor(.x)})
+ names(rearrange_factors) <- factor_names
+
+ # Remove tmp vars and add factors
+ data <- base_deselect_(data, cols = tmp_rearrange_vars) %>%
+ add_dimensions_(new_vectors = rearrange_factors,
+ overwrite = overwrite)
+ } else {
+ # If names are NULL, we just remove the tmp columns
+ data <- base_deselect_(data, cols = tmp_rearrange_vars)
+ }
+
+ data
+}
+
+# Wrapper for running extreme pairing and ordering data by it
+order_and_pair_extremes_ <- function(data, col, new_col, unequal_method){
+
+ # Order data frame
+ data <- data[order(data[[col]]), , drop = FALSE]
+
+ # Add rearrange factor (of type integer)
+ data[[new_col]] <-
+ create_rearrange_factor_pair_extremes_(
+ size = nrow(data),
+ unequal_method = unequal_method
+ )
data
}
diff --git a/R/rearrangers.R b/R/rearrangers.R
index e6e4694..fb702f3 100644
--- a/R/rearrangers.R
+++ b/R/rearrangers.R
@@ -196,6 +196,29 @@ centering_rearranger_ <- function(data,
#' @param shuffle_members Whether to shuffle the pair members. (Logical)
#' @param shuffle_pairs Whether to shuffle the pairs. (Logical)
#' @param factor_name Name of new column with the sorting factor. If \code{NULL}, no column is added.
+#' @param num_pairings Number of pairings to perform (recursively). At least \code{1}.
+#'
+#' Based on \code{`balance`}, the secondary pairings perform extreme pairing on either the
+#' \emph{sum}, \emph{absolute difference}, \emph{min}, or \emph{max} of the pair elements.
+#' @param balance What to balance pairs for in a given \emph{secondary} pairing.
+#' Either \code{"mean"}, \code{"spread"}, \code{"min"}, or \code{"max"}.
+#' Can be a single string used for all secondary pairings
+#' or one for each secondary pairing (\code{`num_pairings` - 1}).
+#'
+#' The first pairing always pairs the actual element values.
+#'
+#' \subsection{mean}{
+#' Pairs have similar means. The values in the pairs from the previous pairing
+#' are aggregated with \code{`sum()`} and paired.
+#' }
+#' \subsection{spread}{
+#' Pairs have similar spread (e.g. standard deviations). The values in the pairs from the previous pairing
+#' are aggregated with \code{`sum(abs(diff()))`} and paired.
+#' }
+#' \subsection{min / max}{
+#' Pairs have similar minimum / maximum values. The values in the pairs from the previous pairing
+#' are aggregated with \code{`min()`} / \code{`max()`} and paired.
+#' }
#' @param unequal_method Method for dealing with an unequal number of rows
#' in \code{`data`}.
#'
@@ -206,7 +229,7 @@ centering_rearranger_ <- function(data,
#'
#' \strong{Example}:
#'
-#' The column values:
+#' The ordered column values:
#'
#' \code{c(1, 2, 3, 4, 5)}
#'
@@ -225,7 +248,7 @@ centering_rearranger_ <- function(data,
#'
#' \strong{Example}:
#'
-#' The column values:
+#' The ordered column values:
#'
#' \code{c(1, 2, 3, 4, 5)}
#'
@@ -243,7 +266,7 @@ centering_rearranger_ <- function(data,
#'
#' \strong{Example}:
#'
-#' The column values:
+#' The ordered column values:
#'
#' \code{c(1, 2, 3, 4, 5)}
#'
@@ -259,9 +282,9 @@ centering_rearranger_ <- function(data,
#' @keywords internal
#' @return
#' The sorted \code{data.frame} (\code{tibble}) / \code{vector}.
-#' Optionally with the sorting factor added.
+#' Optionally with the sorting factor(s) added.
#'
-#' When \code{`data`} is a \code{vector} and \code{`keep_factors`} is \code{FALSE},
+#' When \code{`data`} is a \code{vector} and \code{`factor_name`} is \code{NULL},
#' the output will be a \code{vector}. Otherwise, a \code{data.frame}.
extreme_pairing_rearranger_ <- function(data,
col = NULL,
@@ -269,6 +292,7 @@ extreme_pairing_rearranger_ <- function(data,
shuffle_members = FALSE,
shuffle_pairs = FALSE,
num_pairings = 1,
+ balance = "mean",
factor_name = ".pair",
overwrite = FALSE) {
@@ -276,14 +300,22 @@ extreme_pairing_rearranger_ <- function(data,
assert_collection <- checkmate::makeAssertCollection()
checkmate::assert_count(num_pairings, positive = TRUE, add = assert_collection)
checkmate::assert_string(unequal_method, min.chars = 1, add = assert_collection)
+ checkmate::assert_character(balance, min.chars = 1, any.missing = FALSE, add = assert_collection)
checkmate::assert_string(factor_name, min.chars = 1, null.ok = TRUE, add = assert_collection)
checkmate::assert_flag(shuffle_members, add = assert_collection)
checkmate::assert_flag(shuffle_pairs, add = assert_collection)
checkmate::reportAssertions(assert_collection)
checkmate::assert_names(unequal_method,
- subset.of = c("first", "middle", "last"),
- add = assert_collection
- )
+ subset.of = c("first", "middle", "last"),
+ add = assert_collection)
+ checkmate::assert_names(balance,
+ subset.of = c("mean", "spread", "min", "max"),
+ add = assert_collection)
+ checkmate::reportAssertions(assert_collection)
+ if (num_pairings > 1 &&
+ length(balance) %ni% c(1, num_pairings - 1)){
+ assert_collection$push("length of 'balance' must be either 1 or 'num_pairings' - 1.")
+ }
checkmate::reportAssertions(assert_collection)
# End of argument checks ####
@@ -296,6 +328,7 @@ extreme_pairing_rearranger_ <- function(data,
overwrite = overwrite,
unequal_method = unequal_method,
num_pairings = num_pairings,
+ balance = balance,
shuffle_members = shuffle_members,
shuffle_pairs = shuffle_pairs,
factor_name = factor_name
diff --git a/R/shuffle_hierarchy.R b/R/shuffle_hierarchy.R
new file mode 100644
index 0000000..9bdac7f
--- /dev/null
+++ b/R/shuffle_hierarchy.R
@@ -0,0 +1,133 @@
+
+
+# __________________ #< 563edba79bb23631ec71006e35dbc06a ># __________________
+# Shuffle hierarchy ####
+
+
+#' @title Shuffle multi-column hierarchy of groups
+#' @description
+#' \Sexpr[results=rd, stage=render]{lifecycle::badge("experimental")}
+#'
+#' Shuffles a tree/hierarchy of groups, one column at a time.
+#' The levels in the last ("leaf") column are shuffled first, then the second-last column, and so on.
+#' Elements of the same group are ordered sequentially.
+#' @author Ludvig Renbo Olsen, \email{r-pkgs@@ludvigolsen.dk}
+#' @export
+#' @family rearrange functions
+#' @param data \code{data.frame}.
+#' @param group_cols Names of columns making up the group hierarchy.
+#' The last column is the \emph{leaf} and is shuffled first (if also in \code{`cols_to_shuffle`}).
+#' @param cols_to_shuffle Names of columns to shuffle hierarchically.
+#' By default, all the \code{`group_cols`} are shuffled.
+#' @param leaf_has_groups Whether the leaf column contains groups or values. (Logical)
+#'
+#' When the elements are \emph{group identifiers}, they are ordered sequentially and shuffled together.
+#'
+#' When the elements are \emph{values}, they are simply shuffled.
+#' @return
+#' The shuffled \code{data.frame} (\code{tibble}).
+#' @examples
+#' # Attach packages
+#' library(rearrr)
+#' library(dplyr)
+#'
+#' df <- data.frame(
+#' 'a' = rep(1:4, each = 4),
+#' 'b' = rep(1:8, each = 2),
+#' 'c' = 1:16
+#' )
+#'
+#' # Set seed for reproducibility
+#' set.seed(2)
+#'
+#' # Shuffle all columns
+#' shuffle_hierarchy(df, group_cols = c('a', 'b', 'c'))
+#'
+#' # Don't shuffle 'b' but keep grouping by it
+#' # So 'c' will be shuffled within each group in 'b'
+#' shuffle_hierarchy(
+#' data = df,
+#' group_cols = c('a', 'b', 'c'),
+#' cols_to_shuffle = c('a', 'c')
+#' )
+#'
+#' # Shuffle 'b' as if it's not a group column
+#' # so elements are independent within their group
+#' # (i.e. same-valued elements are not necessarily ordered sequentially)
+#' shuffle_hierarchy(df, group_cols = c('a', 'b'), leaf_has_groups = FALSE)
+shuffle_hierarchy <- function(data, group_cols, cols_to_shuffle = group_cols, leaf_has_groups = TRUE) {
+
+ # Check arguments ####
+ assert_collection <- checkmate::makeAssertCollection()
+ checkmate::assert_data_frame(data, add = assert_collection)
+ checkmate::assert_character(group_cols, min.chars = 1, any.missing = FALSE,
+ unique = TRUE, add = assert_collection)
+ checkmate::assert_character(cols_to_shuffle, min.chars = 1, any.missing = FALSE,
+ unique = TRUE, add = assert_collection)
+ checkmate::assert_flag(leaf_has_groups, add = assert_collection)
+ checkmate::reportAssertions(assert_collection)
+ if (length(setdiff(cols_to_shuffle, group_cols))){
+ assert_collection$push("'cols_to_shuffle' can only contain names that are also in 'group_cols'.")
+ }
+ if (dplyr::is_grouped_df(data)){
+ warning("'data' is already grouped. Those groups will be ignored.")
+ }
+ checkmate::reportAssertions(assert_collection)
+ # End of argument checks ####
+
+ # Extract leaf column
+ leaf_col <- tail(group_cols, 1)
+
+ # Get environment to update 'data' in
+ data_env <- environment()
+
+ # Order 'data' by one column at the time
+ plyr::l_ply(rev(seq_along(group_cols)), function(gc_idx) {
+ if (group_cols[[gc_idx]] %in% cols_to_shuffle) {
+ # Find the grouping columns to apply
+ to_group_by <- head(group_cols, n = gc_idx - 1)
+
+ # Group 'data'
+ data <- dplyr::group_by(data, !!!rlang::syms(to_group_by))
+
+ if (leaf_col == group_cols[[gc_idx]] &&
+ !isTRUE(leaf_has_groups)) {
+ # Shuffle leaf where each element is independent
+ # (not in a group)
+ data <- data %>%
+ dplyr::sample_frac()
+ } else {
+ # Shuffle by unique group levels
+ data <- run_by_group(data,
+ fn = shuffle_uniques_,
+ col = group_cols[[gc_idx]])
+ }
+
+ # Assign to parent environment
+ data_env[["data"]] <- data
+ }
+ })
+
+ # Ungroup and return
+ data %>%
+ dplyr::ungroup()
+
+}
+
+# Extract unique values in the column
+# and sort 'data' by their shuffled index
+shuffle_uniques_ <- function(data, grp_id, col){
+ tmp_index <- create_tmp_var(data)
+
+ # Extract and shuffle unique group levels
+ uniques <- unique(data[[col]]) %>%
+ tibble::enframe(name = NULL, value = col) %>%
+ dplyr::sample_frac() %>%
+ dplyr::mutate(!!tmp_index := dplyr::row_number())
+
+ # Order 'data' by the shuffled group levels
+ data %>%
+ dplyr::left_join(uniques, by = col) %>%
+ dplyr::arrange(!!as.name(tmp_index)) %>%
+ base_deselect_(cols = tmp_index)
+}
diff --git a/README.Rmd b/README.Rmd
index 7a556a6..3f5c415 100644
--- a/README.Rmd
+++ b/README.Rmd
@@ -39,7 +39,7 @@ rvers <- substring(dep, 7, nchar(dep)-1)
[](https://codecov.io/gh/ludvigolsen/rearrr?branch=master)
[](https://travis-ci.com/LudvigOlsen/rearrr)
[](https://ci.appveyor.com/project/LudvigOlsen/rearrr)
-
+[](https://zenodo.org/badge/latestdoi/259158437)
## Overview
@@ -94,6 +94,7 @@ Development version:
|`furthest_from()` |Order values by longest distance to an origin. |
|`rev_windows()` |Reverse order window-wise. |
|`roll_elements()` |Rolls/shifts positions of elements. |
+|`shuffle_hierarchy()` |Shuffle multi-column hierarchy of groups. |
### Mutators
@@ -307,6 +308,22 @@ orderings %>%
scale_colour_brewer(palette = "Dark2")
```
+### Shuffle Hierarchy
+
+When having a `data.frame` with multiple grouping columns, we can shuffle them one column (hierarchical level) at a time:
+
+```{r eval=FALSE}
+# Shuffle a given data frame 'df'
+shuffle_hierarchy(df, group_cols = c("a", "b", "c"))
+```
+
+The columns are shuffled one at a time, as so:
+
+
+
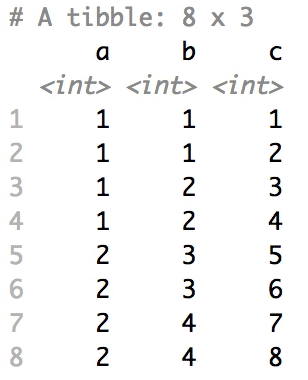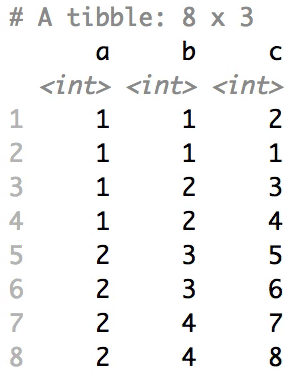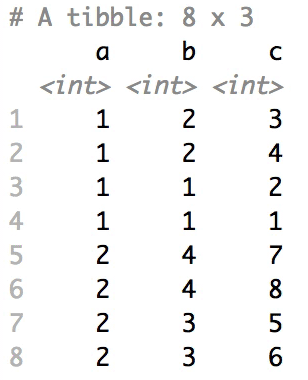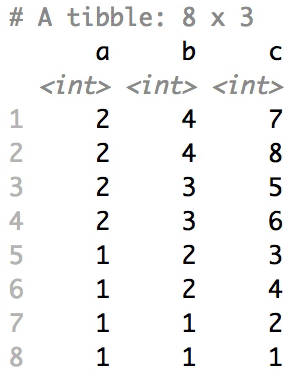
+
 +### Shuffle Hierarchy
+
+When having a `data.frame` with multiple grouping columns, we can
+shuffle them one column (hierarchical level) at a time:
+
+``` r
+# Shuffle a given data frame 'df'
+shuffle_hierarchy(df, group_cols = c("a", "b", "c"))
+```
+
+The columns are shuffled one at a time, as
+so:
+
+
+### Shuffle Hierarchy
+
+When having a `data.frame` with multiple grouping columns, we can
+shuffle them one column (hierarchical level) at a time:
+
+``` r
+# Shuffle a given data frame 'df'
+shuffle_hierarchy(df, group_cols = c("a", "b", "c"))
+```
+
+The columns are shuffled one at a time, as
+so:
+
+
+
+
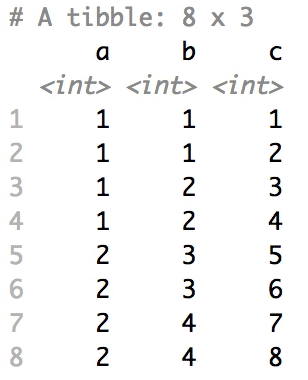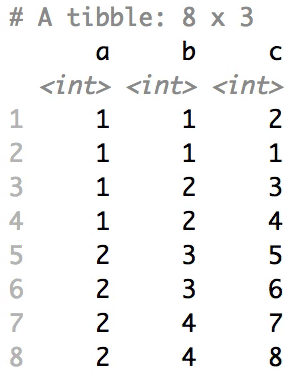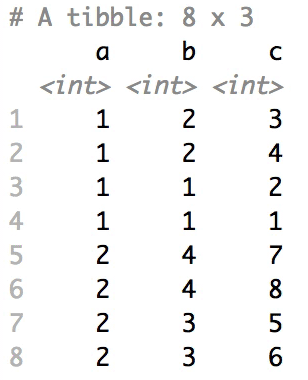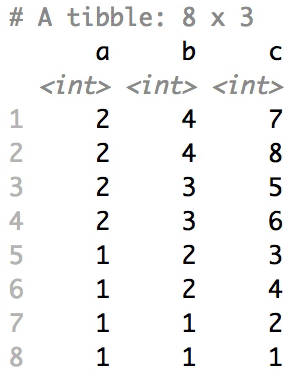
+
+
 +
+ 3-dimensional rotation:
@@ -353,7 +374,7 @@ rotate_3d(
#> 12 12 18 0.505 -1.67 9.88 -5.04
+
3-dimensional rotation:
@@ -353,7 +374,7 @@ rotate_3d(
#> 12 12 18 0.505 -1.67 9.88 -5.04
+ ### Swirl values
@@ -461,7 +482,7 @@ expand_distances(
2d
expansion:
-
### Swirl values
@@ -461,7 +482,7 @@ expand_distances(
2d
expansion:
- +
+ Expand differently in each axis:
@@ -489,7 +510,7 @@ expand_distances_each(
#> 10 0.361 0.169 -0.466 0.251 x=7,y=0.5
```
-
Expand differently in each axis:
@@ -489,7 +510,7 @@ expand_distances_each(
#> 10 0.361 0.169 -0.466 0.251 x=7,y=0.5
```
- +
+ ### Cluster groups
@@ -525,7 +546,7 @@ cluster_groups(
#> # … with 40 more rows
```
-
### Cluster groups
@@ -525,7 +546,7 @@ cluster_groups(
#> # … with 40 more rows
```
- +
+ ### Dim values
@@ -558,7 +579,7 @@ df_clustered %>%
#> # … with 40 more rows
```
-
### Dim values
@@ -558,7 +579,7 @@ df_clustered %>%
#> # … with 40 more rows
```
- +
+ ### Flip values
@@ -587,7 +608,7 @@ flip_values(
#> 10 0.0618 1.14
```
-
### Flip values
@@ -587,7 +608,7 @@ flip_values(
#> 10 0.0618 1.14
```
- +
+ ## Forming examples
@@ -722,7 +743,7 @@ generate_clusters(
#> # … with 40 more rows
```
-
## Forming examples
@@ -722,7 +743,7 @@ generate_clusters(
#> # … with 40 more rows
```
- +
+ ## Utilities
diff --git a/cran-comments.md b/cran-comments.md
index 4b4fe6a..02c18c2 100644
--- a/cran-comments.md
+++ b/cran-comments.md
@@ -5,9 +5,4 @@
## R CMD check results
-0 errors | 0 warnings | 1 note
-
-* This is a new release.
-* Removed examples for unexported functions
-* Made examples testable in most cases
-* Replaced <<- assignment with specific assignment to the right environment
+0 errors | 0 warnings | 0 notes
diff --git a/man/center_max.Rd b/man/center_max.Rd
index 729ce93..0a316f0 100644
--- a/man/center_max.Rd
+++ b/man/center_max.Rd
@@ -84,7 +84,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/center_min.Rd b/man/center_min.Rd
index e965bf7..0131e96 100644
--- a/man/center_min.Rd
+++ b/man/center_min.Rd
@@ -84,7 +84,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/closest_to.Rd b/man/closest_to.Rd
index 3c8a1d1..a585d95 100644
--- a/man/closest_to.Rd
+++ b/man/closest_to.Rd
@@ -165,7 +165,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
Other distance functions:
\code{\link{dim_values}()},
diff --git a/man/extreme_pairing_rearranger_.Rd b/man/extreme_pairing_rearranger_.Rd
index cb83c7e..4ede22f 100644
--- a/man/extreme_pairing_rearranger_.Rd
+++ b/man/extreme_pairing_rearranger_.Rd
@@ -11,6 +11,7 @@ extreme_pairing_rearranger_(
shuffle_members = FALSE,
shuffle_pairs = FALSE,
num_pairings = 1,
+ balance = "mean",
factor_name = ".pair",
overwrite = FALSE
)
@@ -31,7 +32,7 @@ The first group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -50,7 +51,7 @@ The middle group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -68,7 +69,7 @@ The last group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -86,15 +87,40 @@ And are \strong{ordered as}:
\item{shuffle_pairs}{Whether to shuffle the pairs. (Logical)}
+\item{num_pairings}{Number of pairings to perform (recursively). At least \code{1}.
+
+Based on \code{`balance`}, the secondary pairings perform extreme pairing on either the
+\emph{sum}, \emph{absolute difference}, \emph{min}, or \emph{max} of the pair elements.}
+
+\item{balance}{What to balance pairs for in a given \emph{secondary} pairing.
+Either \code{"mean"}, \code{"spread"}, \code{"min"}, or \code{"max"}.
+Can be a single string used for all secondary pairings
+or one for each secondary pairing (\code{`num_pairings` - 1}).
+
+The first pairing always pairs the actual element values.
+
+\subsection{mean}{
+Pairs have similar means. The values in the pairs from the previous pairing
+are aggregated with \code{`sum()`} and paired.
+}
+\subsection{spread}{
+Pairs have similar spread (e.g. standard deviations). The values in the pairs from the previous pairing
+are aggregated with \code{`sum(abs(diff()))`} and paired.
+}
+\subsection{min / max}{
+Pairs have similar minimum / maximum values. The values in the pairs from the previous pairing
+are aggregated with \code{`min()`} / \code{`max()`} and paired.
+}}
+
\item{factor_name}{Name of new column with the sorting factor. If \code{NULL}, no column is added.}
\item{overwrite}{Whether to allow overwriting of existing columns. (Logical)}
}
\value{
The sorted \code{data.frame} (\code{tibble}) / \code{vector}.
-Optionally with the sorting factor added.
+Optionally with the sorting factor(s) added.
-When \code{`data`} is a \code{vector} and \code{`keep_factors`} is \code{FALSE},
+When \code{`data`} is a \code{vector} and \code{`factor_name`} is \code{NULL},
the output will be a \code{vector}. Otherwise, a \code{data.frame}.
}
\description{
diff --git a/man/figures/README-unnamed-chunk-19-1.png b/man/figures/README-unnamed-chunk-19-1.png
new file mode 100644
index 0000000..e6e93ee
Binary files /dev/null and b/man/figures/README-unnamed-chunk-19-1.png differ
diff --git a/man/figures/README-unnamed-chunk-21-1.png b/man/figures/README-unnamed-chunk-21-1.png
new file mode 100644
index 0000000..766574a
Binary files /dev/null and b/man/figures/README-unnamed-chunk-21-1.png differ
diff --git a/man/figures/README-unnamed-chunk-27-1.png b/man/figures/README-unnamed-chunk-27-1.png
new file mode 100644
index 0000000..927a9f9
Binary files /dev/null and b/man/figures/README-unnamed-chunk-27-1.png differ
diff --git a/man/figures/README-unnamed-chunk-29-1.png b/man/figures/README-unnamed-chunk-29-1.png
new file mode 100644
index 0000000..3c5a2ff
Binary files /dev/null and b/man/figures/README-unnamed-chunk-29-1.png differ
diff --git a/man/figures/README-unnamed-chunk-31-1.png b/man/figures/README-unnamed-chunk-31-1.png
new file mode 100644
index 0000000..5e97abd
Binary files /dev/null and b/man/figures/README-unnamed-chunk-31-1.png differ
diff --git a/man/figures/README-unnamed-chunk-33-1.png b/man/figures/README-unnamed-chunk-33-1.png
new file mode 100644
index 0000000..2487964
Binary files /dev/null and b/man/figures/README-unnamed-chunk-33-1.png differ
diff --git a/man/figures/README-unnamed-chunk-35-1.png b/man/figures/README-unnamed-chunk-35-1.png
new file mode 100644
index 0000000..0e4b3d0
Binary files /dev/null and b/man/figures/README-unnamed-chunk-35-1.png differ
diff --git a/man/figures/README-unnamed-chunk-46-1.png b/man/figures/README-unnamed-chunk-46-1.png
new file mode 100644
index 0000000..24b9de6
Binary files /dev/null and b/man/figures/README-unnamed-chunk-46-1.png differ
diff --git a/man/furthest_from.Rd b/man/furthest_from.Rd
index 4f7980a..17afb31 100644
--- a/man/furthest_from.Rd
+++ b/man/furthest_from.Rd
@@ -166,7 +166,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
Other distance functions:
\code{\link{closest_to}()},
diff --git a/man/pair_extremes.Rd b/man/pair_extremes.Rd
index b0eb76b..08fe718 100644
--- a/man/pair_extremes.Rd
+++ b/man/pair_extremes.Rd
@@ -9,15 +9,19 @@ pair_extremes(
data,
col = NULL,
unequal_method = "middle",
+ num_pairings = 1,
+ balance = "mean",
shuffle_members = FALSE,
shuffle_pairs = FALSE,
- factor_name = ".pair",
+ factor_name = ifelse(num_pairings == 1, ".pair", ".pairing"),
overwrite = FALSE
)
pair_extremes_vec(
data,
unequal_method = "middle",
+ num_pairings = 1,
+ balance = "mean",
shuffle_members = FALSE,
shuffle_pairs = FALSE
)
@@ -38,7 +42,7 @@ The first group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -57,7 +61,7 @@ The middle group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -75,7 +79,7 @@ The last group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -89,6 +93,31 @@ And are \strong{ordered as}:
}}
+\item{num_pairings}{Number of pairings to perform (recursively). At least \code{1}.
+
+Based on \code{`balance`}, the secondary pairings perform extreme pairing on either the
+\emph{sum}, \emph{absolute difference}, \emph{min}, or \emph{max} of the pair elements.}
+
+\item{balance}{What to balance pairs for in a given \emph{secondary} pairing.
+Either \code{"mean"}, \code{"spread"}, \code{"min"}, or \code{"max"}.
+Can be a single string used for all secondary pairings
+or one for each secondary pairing (\code{`num_pairings` - 1}).
+
+The first pairing always pairs the actual element values.
+
+\subsection{mean}{
+Pairs have similar means. The values in the pairs from the previous pairing
+are aggregated with \code{`sum()`} and paired.
+}
+\subsection{spread}{
+Pairs have similar spread (e.g. standard deviations). The values in the pairs from the previous pairing
+are aggregated with \code{`sum(abs(diff()))`} and paired.
+}
+\subsection{min / max}{
+Pairs have similar minimum / maximum values. The values in the pairs from the previous pairing
+are aggregated with \code{`min()`} / \code{`max()`} and paired.
+}}
+
\item{shuffle_members}{Whether to shuffle the pair members. (Logical)}
\item{shuffle_pairs}{Whether to shuffle the pairs. (Logical)}
@@ -166,6 +195,9 @@ pair_extremes(df, col = "A", shuffle_members = TRUE)
# Shuffle the order of the pairs
pair_extremes(df, col = "A", shuffle_pairs = TRUE)
+# Use recursive pairing
+pair_extremes(df, col = "A", num_pairings = 2)
+
# Grouped by G
df \%>\%
dplyr::select(G, A) \%>\% # For clarity
@@ -197,7 +229,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/position_max.Rd b/man/position_max.Rd
index 751e6ea..06a8d0c 100644
--- a/man/position_max.Rd
+++ b/man/position_max.Rd
@@ -89,7 +89,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/position_min.Rd b/man/position_min.Rd
index f1e85ae..e12c003 100644
--- a/man/position_min.Rd
+++ b/man/position_min.Rd
@@ -91,7 +91,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_max}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/rev_windows.Rd b/man/rev_windows.Rd
index a81f40b..59d46ae 100644
--- a/man/rev_windows.Rd
+++ b/man/rev_windows.Rd
@@ -91,7 +91,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_max}()},
\code{\link{position_min}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/roll_elements.Rd b/man/roll_elements.Rd
index f4c6bd8..e2ef28f 100644
--- a/man/roll_elements.Rd
+++ b/man/roll_elements.Rd
@@ -137,7 +137,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_max}()},
\code{\link{position_min}()},
-\code{\link{rev_windows}()}
+\code{\link{rev_windows}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/shuffle_hierarchy.Rd b/man/shuffle_hierarchy.Rd
new file mode 100644
index 0000000..dd49c84
--- /dev/null
+++ b/man/shuffle_hierarchy.Rd
@@ -0,0 +1,84 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/shuffle_hierarchy.R
+\name{shuffle_hierarchy}
+\alias{shuffle_hierarchy}
+\title{Shuffle multi-column hierarchy of groups}
+\usage{
+shuffle_hierarchy(
+ data,
+ group_cols,
+ cols_to_shuffle = group_cols,
+ leaf_has_groups = TRUE
+)
+}
+\arguments{
+\item{data}{\code{data.frame}.}
+
+\item{group_cols}{Names of columns making up the group hierarchy.
+The last column is the \emph{leaf} and is shuffled first (if also in \code{`cols_to_shuffle`}).}
+
+\item{cols_to_shuffle}{Names of columns to shuffle hierarchically.
+By default, all the \code{`group_cols`} are shuffled.}
+
+\item{leaf_has_groups}{Whether the leaf column contains groups or values. (Logical)
+
+When the elements are \emph{group identifiers}, they are ordered sequentially and shuffled together.
+
+When the elements are \emph{values}, they are simply shuffled.}
+}
+\value{
+The shuffled \code{data.frame} (\code{tibble}).
+}
+\description{
+\Sexpr[results=rd, stage=render]{lifecycle::badge("experimental")}
+
+Shuffles a tree/hierarchy of groups, one column at a time.
+The levels in the last ("leaf") column are shuffled first, then the second-last column, and so on.
+Elements of the same group are ordered sequentially.
+}
+\examples{
+# Attach packages
+library(rearrr)
+library(dplyr)
+
+df <- data.frame(
+ 'a' = rep(1:4, each = 4),
+ 'b' = rep(1:8, each = 2),
+ 'c' = 1:16
+)
+
+# Set seed for reproducibility
+set.seed(2)
+
+# Shuffle all columns
+shuffle_hierarchy(df, group_cols = c('a', 'b', 'c'))
+
+# Don't shuffle 'b' but keep grouping by it
+# So 'c' will be shuffled within each group in 'b'
+shuffle_hierarchy(
+ data = df,
+ group_cols = c('a', 'b', 'c'),
+ cols_to_shuffle = c('a', 'c')
+)
+
+# Shuffle 'b' as if it's not a group column
+# so elements are independent within their group
+# (i.e. same-valued elements are not necessarily ordered sequentially)
+shuffle_hierarchy(df, group_cols = c('a', 'b'), leaf_has_groups = FALSE)
+}
+\seealso{
+Other rearrange functions:
+\code{\link{center_max}()},
+\code{\link{center_min}()},
+\code{\link{closest_to}()},
+\code{\link{furthest_from}()},
+\code{\link{pair_extremes}()},
+\code{\link{position_max}()},
+\code{\link{position_min}()},
+\code{\link{rev_windows}()},
+\code{\link{roll_elements}()}
+}
+\author{
+Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
+}
+\concept{rearrange functions}
diff --git a/tests/testthat/test_pair_extremes.R b/tests/testthat/test_pair_extremes.R
index dda4aa2..52c915f 100644
--- a/tests/testthat/test_pair_extremes.R
+++ b/tests/testthat/test_pair_extremes.R
@@ -2,7 +2,7 @@ library(rearrr)
context("pair_extremes()")
-test_that("rearrange() with method pair_extremes works", {
+test_that("pair_extremes() works", {
# Create data frame
xpectr::set_test_seed(1)
@@ -70,7 +70,7 @@ test_that("rearrange() with method pair_extremes works", {
expect_equal(df_rearranged$score, c(69, 140, 79, 92, 85, 87))
})
-test_that("rearrange() with method pair_extremes throws expected errors", {
+test_that("pair_extremes() throws expected errors", {
df <- data.frame("a" = c(1,2,3))
## Testing 'rearrange(df, method = "pair_extremes", uneq...' ####
@@ -209,7 +209,9 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# "data" = list(df, head(df, 8), c(1,2,3,4,5,6,7), c(1,2,3,4),
# factor(c(1,2,3,4,5,6,7,1,2,3)), list(1,2,3), NA, 1),
# "col" = list(NULL, "A", "B", "C"),
- # "unequal_method" = list("middle", "first", "last"),
+ # "unequal_method" = list("middle", "first", "last", NA),
+ # "num_pairings" = list(1, 2, NA),
+ # "balance" = list("mean", "spread", NA),
# "shuffle_members" = list(FALSE, TRUE),
# "shuffle_pairs" = list(FALSE, TRUE),
# "factor_name" = list(NULL, ".pair", "A", 1, NA),
@@ -219,7 +221,10 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# list("data" = c(1,2,3,4), "col" = "A"),
# list("data" = c(1,2,3,4), "factor_name" = "another_name"),
# list("shuffle_members" = TRUE, "shuffle_pairs" = TRUE),
- # list("factor_name" = "A", "overwrite" = FALSE)
+ # list("factor_name" = "A", "overwrite" = FALSE),
+ # list("num_pairings" = 2, "balance" = "spread"),
+ # list("num_pairings" = 2, "balance" = "max"),
+ # list("num_pairings" = 2, "balance" = "min")
# ),
# indentation = 2
# )
@@ -232,7 +237,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Testing pair_extremes(data = df, col = NULL, unequal...
xpectr::set_test_seed(42)
# Assigning output
- output_19148 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19148 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19148),
@@ -285,7 +290,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = head(df, 8)
xpectr::set_test_seed(42)
# Assigning output
- output_19370 <- pair_extremes(data = head(df, 8), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19370 <- pair_extremes(data = head(df, 8), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19370),
@@ -338,7 +343,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = c(1, 2, 3, 4, ...
xpectr::set_test_seed(42)
# Assigning output
- output_12861 <- pair_extremes(data = c(1, 2, 3, 4, 5, 6, 7), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_12861 <- pair_extremes(data = c(1, 2, 3, 4, 5, 6, 7), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_12861),
@@ -371,7 +376,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = c(1, 2, 3, 4)
xpectr::set_test_seed(42)
# Assigning output
- output_18304 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_18304 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_18304),
@@ -404,7 +409,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = factor(c(1, 2,...
xpectr::set_test_seed(42)
# Assigning output
- output_16417 <- pair_extremes(data = factor(c(1, 2, 3, 4, 5, 6, 7, 1, 2, 3)), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_16417 <- pair_extremes(data = factor(c(1, 2, 3, 4, 5, 6, 7, 1, 2, 3)), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing is factor
expect_true(
is.factor(output_16417))
@@ -437,7 +442,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_15190 <- xpectr::capture_side_effects(pair_extremes(data = list(1, 2, 3), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_15190 <- xpectr::capture_side_effects(pair_extremes(data = list(1, 2, 3), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_15190[['error']]),
xpectr::strip("1 assertions failed:\n * when 'data' is not a data.frame, it cannot be a list."),
@@ -452,7 +457,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_17365 <- xpectr::capture_side_effects(pair_extremes(data = NA, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_17365 <- xpectr::capture_side_effects(pair_extremes(data = NA, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_17365[['error']]),
xpectr::strip("Assertion failed. One of the following must apply:\n * checkmate::check_data_frame(data): Must be of type 'data.frame', not 'logical'\n * checkmate::check_vector(data): Contains missing values (element 1)\n * checkmate::check_factor(data): Contains missing values (element 1)"),
@@ -466,7 +471,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = 1
xpectr::set_test_seed(42)
# Assigning output
- output_11346 <- pair_extremes(data = 1, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_11346 <- pair_extremes(data = 1, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_11346),
@@ -500,7 +505,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_16569 <- xpectr::capture_side_effects(pair_extremes(data = NULL, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_16569 <- xpectr::capture_side_effects(pair_extremes(data = NULL, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_16569[['error']]),
xpectr::strip("Assertion failed. One of the following must apply:\n * checkmate::check_data_frame(data): Must be of type 'data.frame', not 'NULL'\n * checkmate::check_vector(data): Must be of type 'vector', not 'NULL'\n * checkmate::check_factor(data): Must be of type 'factor', not 'NULL'"),
@@ -515,7 +520,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_17050 <- xpectr::capture_side_effects(pair_extremes(data = c(1, 2, 3, 4), col = "A", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_17050 <- xpectr::capture_side_effects(pair_extremes(data = c(1, 2, 3, 4), col = "A", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_17050[['error']]),
xpectr::strip("1 assertions failed:\n * when 'data' is not a data.frame, 'col(s)' must be 'NULL'."),
@@ -529,7 +534,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data, factor_name
xpectr::set_test_seed(42)
# Assigning output
- output_14577 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "another_name", overwrite = TRUE)
+ output_14577 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "another_name", overwrite = TRUE)
# Testing class
expect_equal(
class(output_14577),
@@ -572,7 +577,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: col = "C"
xpectr::set_test_seed(42)
# Assigning output
- output_17191 <- pair_extremes(data = df, col = "C", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_17191 <- pair_extremes(data = df, col = "C", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_17191),
@@ -625,7 +630,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: col = "A"
xpectr::set_test_seed(42)
# Assigning output
- output_19346 <- pair_extremes(data = df, col = "A", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19346 <- pair_extremes(data = df, col = "A", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19346),
@@ -678,7 +683,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: col = "B"
xpectr::set_test_seed(42)
# Assigning output
- output_12554 <- pair_extremes(data = df, col = "B", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_12554 <- pair_extremes(data = df, col = "B", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_12554),
@@ -731,7 +736,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: unequal_method = "first"
xpectr::set_test_seed(42)
# Assigning output
- output_14622 <- pair_extremes(data = df, col = NULL, unequal_method = "first", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_14622 <- pair_extremes(data = df, col = NULL, unequal_method = "first", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_14622),
@@ -784,7 +789,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: unequal_method = "last"
xpectr::set_test_seed(42)
# Assigning output
- output_19400 <- pair_extremes(data = df, col = NULL, unequal_method = "last", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19400 <- pair_extremes(data = df, col = NULL, unequal_method = "last", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19400),
@@ -834,70 +839,410 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
fixed = TRUE)
# Testing pair_extremes(data = df, col = NULL, unequal...
- # Changed from baseline: unequal_method = NULL
+ # Changed from baseline: unequal_method = NA
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_19782 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = NULL, shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_19782 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = NA, num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_19782[['error']]),
- xpectr::strip("1 assertions failed:\n * Variable 'unequal_method': Must be of type 'string', not 'NULL'."),
+ xpectr::strip("1 assertions failed:\n * Variable 'unequal_method': May not be NA."),
fixed = TRUE)
expect_equal(
xpectr::strip(side_effects_19782[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: unequal_method = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_11174 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = NULL, num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_11174[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'unequal_method': Must be of type 'string', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_11174[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings = 2
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_14749 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_14749),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_14749[["index"]],
+ c(5, 4, 6, 2, 8, 1, 9, 3, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_14749[["A"]],
+ c(4, 5, 7, 8, 1, 9, 2, 3, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_14749[["B"]],
+ c(0.25543, 0.93467, 0.46229, 0.45774, 0.97823, 0.70506, 0.11749,
+ 0.71911, 0.94001),
+ tolerance = 1e-4)
+ expect_equal(
+ output_14749[["C"]],
+ c("E", "D", "F", "B", "H", "A", "I", "C", "G"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_14749),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_14749),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_14749),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_14749),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_14749)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_15603 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = NA, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15603[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'num_pairings': May not be NA."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15603[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_19040 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = NULL, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19040[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'num_pairings': Must be of type 'count', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19040[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings, balance
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_11387 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "spread", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_11387),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_11387[["index"]],
+ c(1, 9, 5, 3, 7, 2, 8, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11387[["A"]],
+ c(9, 2, 4, 3, 6, 8, 1, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11387[["B"]],
+ c(0.70506, 0.11749, 0.25543, 0.71911, 0.94001, 0.45774, 0.97823,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11387[["C"]],
+ c("A", "I", "E", "C", "G", "B", "H", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_11387),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_11387),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_11387),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_11387),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_11387)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings, balance
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19888 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "max", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_19888),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19888[["index"]],
+ c(1, 9, 5, 3, 7, 2, 8, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19888[["A"]],
+ c(9, 2, 4, 3, 6, 8, 1, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19888[["B"]],
+ c(0.70506, 0.11749, 0.25543, 0.71911, 0.94001, 0.45774, 0.97823,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19888[["C"]],
+ c("A", "I", "E", "C", "G", "B", "H", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_19888),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19888),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19888),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19888),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19888)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings, balance
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19466 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "min", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_19466),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19466[["index"]],
+ c(1, 9, 5, 3, 7, 2, 8, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19466[["A"]],
+ c(9, 2, 4, 3, 6, 8, 1, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19466[["B"]],
+ c(0.70506, 0.11749, 0.25543, 0.71911, 0.94001, 0.45774, 0.97823,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19466[["C"]],
+ c("A", "I", "E", "C", "G", "B", "H", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_19466),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19466),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19466),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19466),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19466)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: balance = "spread"
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_10824 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "spread", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_10824),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_10824[["index"]],
+ c(1, 9, 2, 8, 5, 3, 7, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10824[["A"]],
+ c(9, 2, 8, 1, 4, 3, 6, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10824[["B"]],
+ c(0.70506, 0.11749, 0.45774, 0.97823, 0.25543, 0.71911, 0.94001,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10824[["C"]],
+ c("A", "I", "B", "H", "E", "C", "G", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_10824),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_10824),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_10824),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_10824),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_10824)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: balance = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_15142 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = NA, shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15142[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'balance': Contains missing values (element 1)."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15142[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: balance = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_13902 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = NULL, shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_13902[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'balance': Must be of type 'character', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_13902[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
# Testing pair_extremes(data = df, col = NULL, unequal...
# Changed from baseline: shuffle_members = TRUE
xpectr::set_test_seed(42)
# Assigning output
- output_11174 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = TRUE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19057 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = TRUE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
- class(output_11174),
+ class(output_19057),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_11174[["index"]],
- c(9, 1, 8, 2, 5, 3, 7, 6, 4),
+ output_19057[["index"]],
+ c(6, 4, 2, 5, 8, 7, 3, 1, 9),
tolerance = 1e-4)
expect_equal(
- output_11174[["A"]],
- c(2, 9, 1, 8, 4, 3, 6, 7, 5),
+ output_19057[["A"]],
+ c(7, 5, 8, 4, 1, 6, 3, 9, 2),
tolerance = 1e-4)
expect_equal(
- output_11174[["B"]],
- c(0.11749, 0.70506, 0.97823, 0.45774, 0.25543, 0.71911, 0.94001,
- 0.46229, 0.93467),
+ output_19057[["B"]],
+ c(0.46229, 0.93467, 0.45774, 0.25543, 0.97823, 0.94001, 0.71911,
+ 0.70506, 0.11749),
tolerance = 1e-4)
expect_equal(
- output_11174[["C"]],
- c("I", "A", "H", "B", "E", "C", "G", "F", "D"),
+ output_19057[["C"]],
+ c("F", "D", "B", "E", "H", "G", "C", "A", "I"),
fixed = TRUE)
# Testing column names
expect_equal(
- names(output_11174),
+ names(output_19057),
c("index", "A", "B", "C"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_11174),
+ xpectr::element_classes(output_19057),
c("integer", "integer", "numeric", "character"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_11174),
+ xpectr::element_types(output_19057),
c("integer", "integer", "double", "character"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_11174),
+ dim(output_19057),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_11174)),
+ colnames(dplyr::group_keys(output_19057)),
character(0),
fixed = TRUE)
@@ -906,13 +1251,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_14749 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = NULL, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_14469 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = NULL, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_14749[['error']]),
+ xpectr::strip(side_effects_14469[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'shuffle_members': Must be of type 'logical flag', not 'NULL'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_14749[['error_class']]),
+ xpectr::strip(side_effects_14469[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -920,52 +1265,52 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: shuffle_members, shuf...
xpectr::set_test_seed(42)
# Assigning output
- output_15603 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = TRUE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
+ output_18360 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = TRUE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
- class(output_15603),
+ class(output_18360),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_15603[["index"]],
- c(5, 6, 4, 9, 1, 2, 8, 3, 7),
+ output_18360[["index"]],
+ c(7, 3, 2, 8, 5, 4, 6, 9, 1),
tolerance = 1e-4)
expect_equal(
- output_15603[["A"]],
- c(4, 7, 5, 2, 9, 8, 1, 3, 6),
+ output_18360[["A"]],
+ c(6, 3, 8, 1, 4, 5, 7, 2, 9),
tolerance = 1e-4)
expect_equal(
- output_15603[["B"]],
- c(0.25543, 0.46229, 0.93467, 0.11749, 0.70506, 0.45774, 0.97823,
- 0.71911, 0.94001),
+ output_18360[["B"]],
+ c(0.94001, 0.71911, 0.45774, 0.97823, 0.25543, 0.93467, 0.46229,
+ 0.11749, 0.70506),
tolerance = 1e-4)
expect_equal(
- output_15603[["C"]],
- c("E", "F", "D", "I", "A", "B", "H", "C", "G"),
+ output_18360[["C"]],
+ c("G", "C", "B", "H", "E", "D", "F", "I", "A"),
fixed = TRUE)
# Testing column names
expect_equal(
- names(output_15603),
+ names(output_18360),
c("index", "A", "B", "C"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_15603),
+ xpectr::element_classes(output_18360),
c("integer", "integer", "numeric", "character"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_15603),
+ xpectr::element_types(output_18360),
c("integer", "integer", "double", "character"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_15603),
+ dim(output_18360),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_15603)),
+ colnames(dplyr::group_keys(output_18360)),
character(0),
fixed = TRUE)
@@ -973,52 +1318,52 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: shuffle_pairs = TRUE
xpectr::set_test_seed(42)
# Assigning output
- output_19040 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
+ output_17375 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
- class(output_19040),
+ class(output_17375),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_19040[["index"]],
- c(5, 4, 6, 1, 9, 2, 8, 3, 7),
+ output_17375[["index"]],
+ c(9, 1, 2, 8, 5, 7, 3, 4, 6),
tolerance = 1e-4)
expect_equal(
- output_19040[["A"]],
- c(4, 5, 7, 9, 2, 8, 1, 3, 6),
+ output_17375[["A"]],
+ c(2, 9, 8, 1, 4, 6, 3, 5, 7),
tolerance = 1e-4)
expect_equal(
- output_19040[["B"]],
- c(0.25543, 0.93467, 0.46229, 0.70506, 0.11749, 0.45774, 0.97823,
- 0.71911, 0.94001),
+ output_17375[["B"]],
+ c(0.11749, 0.70506, 0.45774, 0.97823, 0.25543, 0.94001, 0.71911,
+ 0.93467, 0.46229),
tolerance = 1e-4)
expect_equal(
- output_19040[["C"]],
- c("E", "D", "F", "A", "I", "B", "H", "C", "G"),
+ output_17375[["C"]],
+ c("I", "A", "B", "H", "E", "G", "C", "D", "F"),
fixed = TRUE)
# Testing column names
expect_equal(
- names(output_19040),
+ names(output_17375),
c("index", "A", "B", "C"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_19040),
+ xpectr::element_classes(output_17375),
c("integer", "integer", "numeric", "character"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_19040),
+ xpectr::element_types(output_17375),
c("integer", "integer", "double", "character"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_19040),
+ dim(output_17375),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_19040)),
+ colnames(dplyr::group_keys(output_17375)),
character(0),
fixed = TRUE)
@@ -1027,13 +1372,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_11387 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = NULL, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_18110 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = NULL, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_11387[['error']]),
+ xpectr::strip(side_effects_18110[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'shuffle_pairs': Must be of type 'logical flag', not 'NULL'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_11387[['error_class']]),
+ xpectr::strip(side_effects_18110[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1041,56 +1386,56 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: factor_name = ".pair"
xpectr::set_test_seed(42)
# Assigning output
- output_19888 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = ".pair", overwrite = TRUE)
+ output_13881 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = ".pair", overwrite = TRUE)
# Testing class
expect_equal(
- class(output_19888),
+ class(output_13881),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_19888[["index"]],
+ output_13881[["index"]],
c(1, 9, 2, 8, 5, 3, 7, 4, 6),
tolerance = 1e-4)
expect_equal(
- output_19888[["A"]],
+ output_13881[["A"]],
c(9, 2, 8, 1, 4, 3, 6, 5, 7),
tolerance = 1e-4)
expect_equal(
- output_19888[["B"]],
+ output_13881[["B"]],
c(0.70506, 0.11749, 0.45774, 0.97823, 0.25543, 0.71911, 0.94001,
0.93467, 0.46229),
tolerance = 1e-4)
expect_equal(
- output_19888[["C"]],
+ output_13881[["C"]],
c("A", "I", "B", "H", "E", "C", "G", "D", "F"),
fixed = TRUE)
expect_equal(
- output_19888[[".pair"]],
+ output_13881[[".pair"]],
structure(c(1L, 1L, 2L, 2L, 3L, 4L, 4L, 5L, 5L), .Label = c("1",
"2", "3", "4", "5"), class = "factor"))
# Testing column names
expect_equal(
- names(output_19888),
+ names(output_13881),
c("index", "A", "B", "C", ".pair"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_19888),
+ xpectr::element_classes(output_13881),
c("integer", "integer", "numeric", "character", "factor"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_19888),
+ xpectr::element_types(output_13881),
c("integer", "integer", "double", "character", "integer"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_19888),
+ dim(output_13881),
c(9L, 5L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_19888)),
+ colnames(dplyr::group_keys(output_13881)),
character(0),
fixed = TRUE)
@@ -1098,52 +1443,52 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: factor_name = "A"
xpectr::set_test_seed(42)
# Assigning output
- output_19466 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = TRUE)
+ output_16851 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = TRUE)
# Testing class
expect_equal(
- class(output_19466),
+ class(output_16851),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_19466[["index"]],
+ output_16851[["index"]],
c(1, 9, 2, 8, 5, 3, 7, 4, 6),
tolerance = 1e-4)
expect_equal(
- output_19466[["A"]],
- structure(c(1L, 1L, 2L, 2L, 3L, 4L, 4L, 5L, 5L), .Label = c("1",
- "2", "3", "4", "5"), class = "factor"))
- expect_equal(
- output_19466[["B"]],
+ output_16851[["B"]],
c(0.70506, 0.11749, 0.45774, 0.97823, 0.25543, 0.71911, 0.94001,
0.93467, 0.46229),
tolerance = 1e-4)
expect_equal(
- output_19466[["C"]],
+ output_16851[["C"]],
c("A", "I", "B", "H", "E", "C", "G", "D", "F"),
fixed = TRUE)
+ expect_equal(
+ output_16851[["A"]],
+ structure(c(1L, 1L, 2L, 2L, 3L, 4L, 4L, 5L, 5L), .Label = c("1",
+ "2", "3", "4", "5"), class = "factor"))
# Testing column names
expect_equal(
- names(output_19466),
- c("index", "A", "B", "C"),
+ names(output_16851),
+ c("index", "B", "C", "A"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_19466),
- c("integer", "factor", "numeric", "character"),
+ xpectr::element_classes(output_16851),
+ c("integer", "numeric", "character", "factor"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_19466),
- c("integer", "integer", "double", "character"),
+ xpectr::element_types(output_16851),
+ c("integer", "double", "character", "integer"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_19466),
+ dim(output_16851),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_19466)),
+ colnames(dplyr::group_keys(output_16851)),
character(0),
fixed = TRUE)
@@ -1152,13 +1497,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_10824 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = 1, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_10039 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = 1, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_10824[['error']]),
+ xpectr::strip(side_effects_10039[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'factor_name': Must be of type 'string' (or 'NULL'), not 'double'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_10824[['error_class']]),
+ xpectr::strip(side_effects_10039[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1167,13 +1512,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_15142 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NA, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_18329 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NA, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_15142[['error']]),
+ xpectr::strip(side_effects_18329[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'factor_name': May not be NA."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_15142[['error_class']]),
+ xpectr::strip(side_effects_18329[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1182,13 +1527,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_13902 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = FALSE), reset_seed = TRUE)
+ side_effects_10073 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = FALSE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_13902[['error']]),
- xpectr::strip("1 assertions failed:\n * The column 'A' already exists and 'overwrite' is disabled."),
+ xpectr::strip(side_effects_10073[['error']]),
+ xpectr::strip("Adding these dimensions would overwrite existing columns: A."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_13902[['error_class']]),
+ xpectr::strip(side_effects_10073[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1197,13 +1542,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_19057 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = NULL), reset_seed = TRUE)
+ side_effects_12076 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = NULL), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_19057[['error']]),
+ xpectr::strip(side_effects_12076[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'overwrite': Must be of type 'logical flag', not 'NULL'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_19057[['error_class']]),
+ xpectr::strip(side_effects_12076[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1233,8 +1578,7 @@ test_that("testing pair_extremes_vec()", {
# Testing values
expect_equal(
output_19148,
- c(0.64175, 0.83045, 0.28614, 0.93708, 0.91481, 0.5191, 0.65699,
- 0.73659, 0.13467),
+ c(0.91480, 0.5190959, 0.736588, 0.65699, 0.6417, 0.8304476, 0.134667, 0.2861395, 0.937075),
tolerance = 1e-4)
# Testing names
expect_equal(
@@ -1252,4 +1596,43 @@ test_that("testing pair_extremes_vec()", {
## Finished testing 'pair_extremes_vec(data = runif(9), unequal_m...' ####
+
+
+ ## Testing 'pair_extremes_vec(data = runif(9), unequal_m...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_16932 <- pair_extremes_vec(data = runif(9),
+ unequal_method = "first",
+ num_pairings = 2)
+ # Testing class
+ expect_equal(
+ class(output_16932),
+ "numeric",
+ fixed = TRUE)
+ # Testing type
+ expect_type(
+ output_16932,
+ type = "double")
+ # Testing values
+ expect_equal(
+ output_16932,
+ c(0.13467, 0.28614, 0.93708, 0.64175, 0.83045, 0.5191, 0.91481,
+ 0.65699, 0.73659),
+ tolerance = 1e-4)
+ # Testing names
+ expect_equal(
+ names(output_16932),
+ NULL,
+ fixed = TRUE)
+ # Testing length
+ expect_equal(
+ length(output_16932),
+ 9L)
+ # Testing sum of element lengths
+ expect_equal(
+ sum(xpectr::element_lengths(output_16932)),
+ 9L)
+ ## Finished testing 'pair_extremes_vec(data = runif(9), unequal_m...' ####
+
})
diff --git a/tests/testthat/test_shuffle_hierarchy.R b/tests/testthat/test_shuffle_hierarchy.R
new file mode 100644
index 0000000..50d36bb
--- /dev/null
+++ b/tests/testthat/test_shuffle_hierarchy.R
@@ -0,0 +1,593 @@
+library(rearrr)
+context("shuffle_hierarchy")
+
+
+test_that("shuffle_hierarchy()", {
+ xpectr::set_test_seed(42)
+
+ df <- data.frame(
+ 'a' = rep(1:2, each = 8),
+ 'b' = rep(1:4, each = 4),
+ 'c' = rep(1:8, each = 2),
+ 'd' = 1:16
+ )
+
+ ## Testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(98)
+ # Assigning output
+ output_19148 <- shuffle_hierarchy(df, group_cols = c('a', 'b', 'c', 'd'))
+ # Testing class
+ expect_equal(
+ class(output_19148),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19148[["a"]],
+ c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["b"]],
+ c(4L, 4L, 4L, 4L, 3L, 3L, 3L, 3L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["c"]],
+ c(8L, 8L, 7L, 7L, 6L, 6L, 5L, 5L, 1L, 1L, 2L, 2L, 4L, 4L, 3L, 3L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["d"]],
+ c(15L, 16L, 13L, 14L, 12L, 11L, 10L, 9L, 1L, 2L, 3L, 4L, 7L, 8L, 5L, 6L),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_19148),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19148),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19148)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+
+
+
+ ## Testing 'shuffle_hierarchy( data = df, group_cols = c...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(98)
+ # Assigning output
+ output_16988 <- shuffle_hierarchy(
+ data = df,
+ group_cols = c('a', 'b', 'c', 'd'),
+ cols_to_shuffle = c('a', 'c')
+ )
+ # Testing class
+ expect_equal(
+ class(output_16988),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_16988[["a"]],
+ c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_16988[["b"]],
+ c(3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_16988[["c"]],
+ c(5L, 5L, 6L, 6L, 7L, 7L, 8L, 8L, 1L, 1L, 2L, 2L, 3L, 3L, 4L, 4L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_16988[["d"]],
+ c(9L, 10L, 11L, 12L, 13L, 14L, 15L, 16L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_16988),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_16988),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_16988),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_16988),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_16988)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy( data = df, group_cols = c...' ####
+
+ ## Testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_10268 <- shuffle_hierarchy(df, group_cols = c('a', 'b'), leaf_has_groups = FALSE)
+ # Testing class
+ expect_equal(
+ class(output_10268),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_10268[["a"]],
+ c(2, 2, 2, 2, 2, 2, 2, 2, 1, 1, 1, 1, 1, 1, 1, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10268[["b"]],
+ c(4, 4, 3, 3, 4, 3, 4, 3, 2, 2, 1, 2, 1, 2, 1, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10268[["c"]],
+ c(7, 7, 6, 6, 8, 5, 8, 5, 4, 4, 1, 3, 2, 3, 2, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10268[["d"]],
+ c(14, 13, 11, 12, 15, 9, 16, 10, 8, 7, 2, 5, 3, 6, 4, 1),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_10268),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_10268),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_10268),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_10268),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_10268)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+
+ # If leaf col is not in cols_to_shuffle
+ # last column should be considered a group (because it is not the leaf)
+
+ ## Testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19782 <- shuffle_hierarchy(df, group_cols = c('a', 'b'), cols_to_shuffle = 'a', leaf_has_groups = FALSE)
+ # Testing class
+ expect_equal(
+ class(output_19782),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19782[["a"]],
+ c(2, 2, 2, 2, 2, 2, 2, 2, 1, 1, 1, 1, 1, 1, 1, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19782[["b"]],
+ c(3, 3, 3, 3, 4, 4, 4, 4, 1, 1, 1, 1, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19782[["c"]],
+ c(5, 5, 6, 6, 7, 7, 8, 8, 1, 1, 2, 2, 3, 3, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19782[["d"]],
+ c(9, 10, 11, 12, 13, 14, 15, 16, 1, 2, 3, 4, 5, 6, 7, 8),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_19782),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19782),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19782),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19782),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19782)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+
+})
+
+
+test_that("fuzz testing shuffle_hierarchy()", {
+ xpectr::set_test_seed(42)
+
+ df <- data.frame(
+ 'a' = rep(1:2, each = 8),
+ 'b' = rep(1:4, each = 4),
+ 'c' = rep(1:8, each = 2),
+ 'd' = 1:16
+ )
+
+ # Generate expectations for 'shuffle_hierarchy'
+ # Tip: comment out the gxs_function() call
+ # so it is easy to regenerate the tests
+ xpectr::set_test_seed(42)
+ # xpectr::gxs_function(
+ # fn = shuffle_hierarchy,
+ # args_values = list(
+ # "data" = list(df, c(1,2,3), NA),
+ # "group_cols" = list(c('a', 'b', 'c', 'd'), c(1, 2, 3, 4), NA),
+ # "cols_to_shuffle" = list(c('a', 'b', 'c', 'd'), c('b', 'c'), 2, NA),
+ # "leaf_has_groups" = list(TRUE, FALSE, NA)
+ # ),
+ # indentation = 2,
+ # copy_env = FALSE
+ # )
+
+ ## Testing 'shuffle_hierarchy' ####
+ ## Initially generated by xpectr
+ # Testing different combinations of argument values
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19148 <- shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_19148),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19148[["a"]],
+ c(1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["b"]],
+ c(1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["c"]],
+ c(2, 2, 1, 1, 3, 3, 4, 4, 6, 6, 5, 5, 8, 8, 7, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["d"]],
+ c(3, 4, 2, 1, 6, 5, 8, 7, 11, 12, 10, 9, 15, 16, 14, 13),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_19148),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19148),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19148)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = c(1, 2, 3), group_c...
+ # Changed from baseline: data = c(1, 2, 3)
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_19370 <- xpectr::capture_side_effects(shuffle_hierarchy(data = c(1, 2, 3), group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19370[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'data': Must be of type 'data.frame', not 'double'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19370[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = NA, group_cols = c(...
+ # Changed from baseline: data = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_12861 <- xpectr::capture_side_effects(shuffle_hierarchy(data = NA, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12861[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'data': Must be of type 'data.frame', not 'logical'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12861[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = NULL, group_cols = ...
+ # Changed from baseline: data = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_18304 <- xpectr::capture_side_effects(shuffle_hierarchy(data = NULL, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_18304[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'data': Must be of type 'data.frame', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_18304[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: group_cols = c(1, 2, ...
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_16417 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c(1, 2, 3, 4), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16417[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'group_cols': Must be of type 'character', not 'double'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16417[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = NA...
+ # Changed from baseline: group_cols = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_15190 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = NA, cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15190[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'group_cols': Contains missing values (element 1)."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15190[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = NU...
+ # Changed from baseline: group_cols = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_17365 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = NULL, cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17365[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'group_cols': Must be of type 'character', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17365[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = c("...
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_11346 <- shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("b", "c"), leaf_has_groups = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_11346),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_11346[["a"]],
+ c(1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11346[["b"]],
+ c(2, 2, 2, 2, 1, 1, 1, 1, 3, 3, 3, 3, 4, 4, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11346[["c"]],
+ c(3, 3, 4, 4, 2, 2, 1, 1, 6, 6, 5, 5, 8, 8, 7, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11346[["d"]],
+ c(5, 6, 7, 8, 3, 4, 1, 2, 11, 12, 9, 10, 15, 16, 13, 14),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_11346),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_11346),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_11346),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_11346),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_11346)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = 2
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_16569 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = 2, leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16569[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'cols_to_shuffle': Must be of type 'character', not 'double'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16569[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_17050 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = NA, leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17050[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'cols_to_shuffle': Contains missing values (element 1)."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17050[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_14577 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = NULL, leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_14577[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'cols_to_shuffle': Must be of type 'character', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_14577[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: leaf_has_groups = FALSE
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_17191 <- shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = FALSE)
+ # Testing class
+ expect_equal(
+ class(output_17191),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_17191[["a"]],
+ c(1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_17191[["b"]],
+ c(1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_17191[["c"]],
+ c(2, 2, 1, 1, 3, 3, 4, 4, 6, 6, 5, 5, 8, 8, 7, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_17191[["d"]],
+ c(3, 4, 2, 1, 6, 5, 8, 7, 11, 12, 10, 9, 15, 16, 14, 13),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_17191),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_17191),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_17191),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_17191),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_17191)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: leaf_has_groups = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_19346 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = NA), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19346[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'leaf_has_groups': May not be NA."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19346[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: leaf_has_groups = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_12554 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = NULL), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12554[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'leaf_has_groups': Must be of type 'logical flag', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12554[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ ## Finished testing 'shuffle_hierarchy' ####
+ #
+
+})
+
## Utilities
diff --git a/cran-comments.md b/cran-comments.md
index 4b4fe6a..02c18c2 100644
--- a/cran-comments.md
+++ b/cran-comments.md
@@ -5,9 +5,4 @@
## R CMD check results
-0 errors | 0 warnings | 1 note
-
-* This is a new release.
-* Removed examples for unexported functions
-* Made examples testable in most cases
-* Replaced <<- assignment with specific assignment to the right environment
+0 errors | 0 warnings | 0 notes
diff --git a/man/center_max.Rd b/man/center_max.Rd
index 729ce93..0a316f0 100644
--- a/man/center_max.Rd
+++ b/man/center_max.Rd
@@ -84,7 +84,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/center_min.Rd b/man/center_min.Rd
index e965bf7..0131e96 100644
--- a/man/center_min.Rd
+++ b/man/center_min.Rd
@@ -84,7 +84,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/closest_to.Rd b/man/closest_to.Rd
index 3c8a1d1..a585d95 100644
--- a/man/closest_to.Rd
+++ b/man/closest_to.Rd
@@ -165,7 +165,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
Other distance functions:
\code{\link{dim_values}()},
diff --git a/man/extreme_pairing_rearranger_.Rd b/man/extreme_pairing_rearranger_.Rd
index cb83c7e..4ede22f 100644
--- a/man/extreme_pairing_rearranger_.Rd
+++ b/man/extreme_pairing_rearranger_.Rd
@@ -11,6 +11,7 @@ extreme_pairing_rearranger_(
shuffle_members = FALSE,
shuffle_pairs = FALSE,
num_pairings = 1,
+ balance = "mean",
factor_name = ".pair",
overwrite = FALSE
)
@@ -31,7 +32,7 @@ The first group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -50,7 +51,7 @@ The middle group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -68,7 +69,7 @@ The last group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -86,15 +87,40 @@ And are \strong{ordered as}:
\item{shuffle_pairs}{Whether to shuffle the pairs. (Logical)}
+\item{num_pairings}{Number of pairings to perform (recursively). At least \code{1}.
+
+Based on \code{`balance`}, the secondary pairings perform extreme pairing on either the
+\emph{sum}, \emph{absolute difference}, \emph{min}, or \emph{max} of the pair elements.}
+
+\item{balance}{What to balance pairs for in a given \emph{secondary} pairing.
+Either \code{"mean"}, \code{"spread"}, \code{"min"}, or \code{"max"}.
+Can be a single string used for all secondary pairings
+or one for each secondary pairing (\code{`num_pairings` - 1}).
+
+The first pairing always pairs the actual element values.
+
+\subsection{mean}{
+Pairs have similar means. The values in the pairs from the previous pairing
+are aggregated with \code{`sum()`} and paired.
+}
+\subsection{spread}{
+Pairs have similar spread (e.g. standard deviations). The values in the pairs from the previous pairing
+are aggregated with \code{`sum(abs(diff()))`} and paired.
+}
+\subsection{min / max}{
+Pairs have similar minimum / maximum values. The values in the pairs from the previous pairing
+are aggregated with \code{`min()`} / \code{`max()`} and paired.
+}}
+
\item{factor_name}{Name of new column with the sorting factor. If \code{NULL}, no column is added.}
\item{overwrite}{Whether to allow overwriting of existing columns. (Logical)}
}
\value{
The sorted \code{data.frame} (\code{tibble}) / \code{vector}.
-Optionally with the sorting factor added.
+Optionally with the sorting factor(s) added.
-When \code{`data`} is a \code{vector} and \code{`keep_factors`} is \code{FALSE},
+When \code{`data`} is a \code{vector} and \code{`factor_name`} is \code{NULL},
the output will be a \code{vector}. Otherwise, a \code{data.frame}.
}
\description{
diff --git a/man/figures/README-unnamed-chunk-19-1.png b/man/figures/README-unnamed-chunk-19-1.png
new file mode 100644
index 0000000..e6e93ee
Binary files /dev/null and b/man/figures/README-unnamed-chunk-19-1.png differ
diff --git a/man/figures/README-unnamed-chunk-21-1.png b/man/figures/README-unnamed-chunk-21-1.png
new file mode 100644
index 0000000..766574a
Binary files /dev/null and b/man/figures/README-unnamed-chunk-21-1.png differ
diff --git a/man/figures/README-unnamed-chunk-27-1.png b/man/figures/README-unnamed-chunk-27-1.png
new file mode 100644
index 0000000..927a9f9
Binary files /dev/null and b/man/figures/README-unnamed-chunk-27-1.png differ
diff --git a/man/figures/README-unnamed-chunk-29-1.png b/man/figures/README-unnamed-chunk-29-1.png
new file mode 100644
index 0000000..3c5a2ff
Binary files /dev/null and b/man/figures/README-unnamed-chunk-29-1.png differ
diff --git a/man/figures/README-unnamed-chunk-31-1.png b/man/figures/README-unnamed-chunk-31-1.png
new file mode 100644
index 0000000..5e97abd
Binary files /dev/null and b/man/figures/README-unnamed-chunk-31-1.png differ
diff --git a/man/figures/README-unnamed-chunk-33-1.png b/man/figures/README-unnamed-chunk-33-1.png
new file mode 100644
index 0000000..2487964
Binary files /dev/null and b/man/figures/README-unnamed-chunk-33-1.png differ
diff --git a/man/figures/README-unnamed-chunk-35-1.png b/man/figures/README-unnamed-chunk-35-1.png
new file mode 100644
index 0000000..0e4b3d0
Binary files /dev/null and b/man/figures/README-unnamed-chunk-35-1.png differ
diff --git a/man/figures/README-unnamed-chunk-46-1.png b/man/figures/README-unnamed-chunk-46-1.png
new file mode 100644
index 0000000..24b9de6
Binary files /dev/null and b/man/figures/README-unnamed-chunk-46-1.png differ
diff --git a/man/furthest_from.Rd b/man/furthest_from.Rd
index 4f7980a..17afb31 100644
--- a/man/furthest_from.Rd
+++ b/man/furthest_from.Rd
@@ -166,7 +166,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
Other distance functions:
\code{\link{closest_to}()},
diff --git a/man/pair_extremes.Rd b/man/pair_extremes.Rd
index b0eb76b..08fe718 100644
--- a/man/pair_extremes.Rd
+++ b/man/pair_extremes.Rd
@@ -9,15 +9,19 @@ pair_extremes(
data,
col = NULL,
unequal_method = "middle",
+ num_pairings = 1,
+ balance = "mean",
shuffle_members = FALSE,
shuffle_pairs = FALSE,
- factor_name = ".pair",
+ factor_name = ifelse(num_pairings == 1, ".pair", ".pairing"),
overwrite = FALSE
)
pair_extremes_vec(
data,
unequal_method = "middle",
+ num_pairings = 1,
+ balance = "mean",
shuffle_members = FALSE,
shuffle_pairs = FALSE
)
@@ -38,7 +42,7 @@ The first group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -57,7 +61,7 @@ The middle group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -75,7 +79,7 @@ The last group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -89,6 +93,31 @@ And are \strong{ordered as}:
}}
+\item{num_pairings}{Number of pairings to perform (recursively). At least \code{1}.
+
+Based on \code{`balance`}, the secondary pairings perform extreme pairing on either the
+\emph{sum}, \emph{absolute difference}, \emph{min}, or \emph{max} of the pair elements.}
+
+\item{balance}{What to balance pairs for in a given \emph{secondary} pairing.
+Either \code{"mean"}, \code{"spread"}, \code{"min"}, or \code{"max"}.
+Can be a single string used for all secondary pairings
+or one for each secondary pairing (\code{`num_pairings` - 1}).
+
+The first pairing always pairs the actual element values.
+
+\subsection{mean}{
+Pairs have similar means. The values in the pairs from the previous pairing
+are aggregated with \code{`sum()`} and paired.
+}
+\subsection{spread}{
+Pairs have similar spread (e.g. standard deviations). The values in the pairs from the previous pairing
+are aggregated with \code{`sum(abs(diff()))`} and paired.
+}
+\subsection{min / max}{
+Pairs have similar minimum / maximum values. The values in the pairs from the previous pairing
+are aggregated with \code{`min()`} / \code{`max()`} and paired.
+}}
+
\item{shuffle_members}{Whether to shuffle the pair members. (Logical)}
\item{shuffle_pairs}{Whether to shuffle the pairs. (Logical)}
@@ -166,6 +195,9 @@ pair_extremes(df, col = "A", shuffle_members = TRUE)
# Shuffle the order of the pairs
pair_extremes(df, col = "A", shuffle_pairs = TRUE)
+# Use recursive pairing
+pair_extremes(df, col = "A", num_pairings = 2)
+
# Grouped by G
df \%>\%
dplyr::select(G, A) \%>\% # For clarity
@@ -197,7 +229,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/position_max.Rd b/man/position_max.Rd
index 751e6ea..06a8d0c 100644
--- a/man/position_max.Rd
+++ b/man/position_max.Rd
@@ -89,7 +89,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/position_min.Rd b/man/position_min.Rd
index f1e85ae..e12c003 100644
--- a/man/position_min.Rd
+++ b/man/position_min.Rd
@@ -91,7 +91,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_max}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/rev_windows.Rd b/man/rev_windows.Rd
index a81f40b..59d46ae 100644
--- a/man/rev_windows.Rd
+++ b/man/rev_windows.Rd
@@ -91,7 +91,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_max}()},
\code{\link{position_min}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/roll_elements.Rd b/man/roll_elements.Rd
index f4c6bd8..e2ef28f 100644
--- a/man/roll_elements.Rd
+++ b/man/roll_elements.Rd
@@ -137,7 +137,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_max}()},
\code{\link{position_min}()},
-\code{\link{rev_windows}()}
+\code{\link{rev_windows}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/shuffle_hierarchy.Rd b/man/shuffle_hierarchy.Rd
new file mode 100644
index 0000000..dd49c84
--- /dev/null
+++ b/man/shuffle_hierarchy.Rd
@@ -0,0 +1,84 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/shuffle_hierarchy.R
+\name{shuffle_hierarchy}
+\alias{shuffle_hierarchy}
+\title{Shuffle multi-column hierarchy of groups}
+\usage{
+shuffle_hierarchy(
+ data,
+ group_cols,
+ cols_to_shuffle = group_cols,
+ leaf_has_groups = TRUE
+)
+}
+\arguments{
+\item{data}{\code{data.frame}.}
+
+\item{group_cols}{Names of columns making up the group hierarchy.
+The last column is the \emph{leaf} and is shuffled first (if also in \code{`cols_to_shuffle`}).}
+
+\item{cols_to_shuffle}{Names of columns to shuffle hierarchically.
+By default, all the \code{`group_cols`} are shuffled.}
+
+\item{leaf_has_groups}{Whether the leaf column contains groups or values. (Logical)
+
+When the elements are \emph{group identifiers}, they are ordered sequentially and shuffled together.
+
+When the elements are \emph{values}, they are simply shuffled.}
+}
+\value{
+The shuffled \code{data.frame} (\code{tibble}).
+}
+\description{
+\Sexpr[results=rd, stage=render]{lifecycle::badge("experimental")}
+
+Shuffles a tree/hierarchy of groups, one column at a time.
+The levels in the last ("leaf") column are shuffled first, then the second-last column, and so on.
+Elements of the same group are ordered sequentially.
+}
+\examples{
+# Attach packages
+library(rearrr)
+library(dplyr)
+
+df <- data.frame(
+ 'a' = rep(1:4, each = 4),
+ 'b' = rep(1:8, each = 2),
+ 'c' = 1:16
+)
+
+# Set seed for reproducibility
+set.seed(2)
+
+# Shuffle all columns
+shuffle_hierarchy(df, group_cols = c('a', 'b', 'c'))
+
+# Don't shuffle 'b' but keep grouping by it
+# So 'c' will be shuffled within each group in 'b'
+shuffle_hierarchy(
+ data = df,
+ group_cols = c('a', 'b', 'c'),
+ cols_to_shuffle = c('a', 'c')
+)
+
+# Shuffle 'b' as if it's not a group column
+# so elements are independent within their group
+# (i.e. same-valued elements are not necessarily ordered sequentially)
+shuffle_hierarchy(df, group_cols = c('a', 'b'), leaf_has_groups = FALSE)
+}
+\seealso{
+Other rearrange functions:
+\code{\link{center_max}()},
+\code{\link{center_min}()},
+\code{\link{closest_to}()},
+\code{\link{furthest_from}()},
+\code{\link{pair_extremes}()},
+\code{\link{position_max}()},
+\code{\link{position_min}()},
+\code{\link{rev_windows}()},
+\code{\link{roll_elements}()}
+}
+\author{
+Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
+}
+\concept{rearrange functions}
diff --git a/tests/testthat/test_pair_extremes.R b/tests/testthat/test_pair_extremes.R
index dda4aa2..52c915f 100644
--- a/tests/testthat/test_pair_extremes.R
+++ b/tests/testthat/test_pair_extremes.R
@@ -2,7 +2,7 @@ library(rearrr)
context("pair_extremes()")
-test_that("rearrange() with method pair_extremes works", {
+test_that("pair_extremes() works", {
# Create data frame
xpectr::set_test_seed(1)
@@ -70,7 +70,7 @@ test_that("rearrange() with method pair_extremes works", {
expect_equal(df_rearranged$score, c(69, 140, 79, 92, 85, 87))
})
-test_that("rearrange() with method pair_extremes throws expected errors", {
+test_that("pair_extremes() throws expected errors", {
df <- data.frame("a" = c(1,2,3))
## Testing 'rearrange(df, method = "pair_extremes", uneq...' ####
@@ -209,7 +209,9 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# "data" = list(df, head(df, 8), c(1,2,3,4,5,6,7), c(1,2,3,4),
# factor(c(1,2,3,4,5,6,7,1,2,3)), list(1,2,3), NA, 1),
# "col" = list(NULL, "A", "B", "C"),
- # "unequal_method" = list("middle", "first", "last"),
+ # "unequal_method" = list("middle", "first", "last", NA),
+ # "num_pairings" = list(1, 2, NA),
+ # "balance" = list("mean", "spread", NA),
# "shuffle_members" = list(FALSE, TRUE),
# "shuffle_pairs" = list(FALSE, TRUE),
# "factor_name" = list(NULL, ".pair", "A", 1, NA),
@@ -219,7 +221,10 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# list("data" = c(1,2,3,4), "col" = "A"),
# list("data" = c(1,2,3,4), "factor_name" = "another_name"),
# list("shuffle_members" = TRUE, "shuffle_pairs" = TRUE),
- # list("factor_name" = "A", "overwrite" = FALSE)
+ # list("factor_name" = "A", "overwrite" = FALSE),
+ # list("num_pairings" = 2, "balance" = "spread"),
+ # list("num_pairings" = 2, "balance" = "max"),
+ # list("num_pairings" = 2, "balance" = "min")
# ),
# indentation = 2
# )
@@ -232,7 +237,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Testing pair_extremes(data = df, col = NULL, unequal...
xpectr::set_test_seed(42)
# Assigning output
- output_19148 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19148 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19148),
@@ -285,7 +290,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = head(df, 8)
xpectr::set_test_seed(42)
# Assigning output
- output_19370 <- pair_extremes(data = head(df, 8), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19370 <- pair_extremes(data = head(df, 8), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19370),
@@ -338,7 +343,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = c(1, 2, 3, 4, ...
xpectr::set_test_seed(42)
# Assigning output
- output_12861 <- pair_extremes(data = c(1, 2, 3, 4, 5, 6, 7), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_12861 <- pair_extremes(data = c(1, 2, 3, 4, 5, 6, 7), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_12861),
@@ -371,7 +376,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = c(1, 2, 3, 4)
xpectr::set_test_seed(42)
# Assigning output
- output_18304 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_18304 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_18304),
@@ -404,7 +409,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = factor(c(1, 2,...
xpectr::set_test_seed(42)
# Assigning output
- output_16417 <- pair_extremes(data = factor(c(1, 2, 3, 4, 5, 6, 7, 1, 2, 3)), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_16417 <- pair_extremes(data = factor(c(1, 2, 3, 4, 5, 6, 7, 1, 2, 3)), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing is factor
expect_true(
is.factor(output_16417))
@@ -437,7 +442,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_15190 <- xpectr::capture_side_effects(pair_extremes(data = list(1, 2, 3), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_15190 <- xpectr::capture_side_effects(pair_extremes(data = list(1, 2, 3), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_15190[['error']]),
xpectr::strip("1 assertions failed:\n * when 'data' is not a data.frame, it cannot be a list."),
@@ -452,7 +457,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_17365 <- xpectr::capture_side_effects(pair_extremes(data = NA, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_17365 <- xpectr::capture_side_effects(pair_extremes(data = NA, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_17365[['error']]),
xpectr::strip("Assertion failed. One of the following must apply:\n * checkmate::check_data_frame(data): Must be of type 'data.frame', not 'logical'\n * checkmate::check_vector(data): Contains missing values (element 1)\n * checkmate::check_factor(data): Contains missing values (element 1)"),
@@ -466,7 +471,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = 1
xpectr::set_test_seed(42)
# Assigning output
- output_11346 <- pair_extremes(data = 1, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_11346 <- pair_extremes(data = 1, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_11346),
@@ -500,7 +505,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_16569 <- xpectr::capture_side_effects(pair_extremes(data = NULL, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_16569 <- xpectr::capture_side_effects(pair_extremes(data = NULL, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_16569[['error']]),
xpectr::strip("Assertion failed. One of the following must apply:\n * checkmate::check_data_frame(data): Must be of type 'data.frame', not 'NULL'\n * checkmate::check_vector(data): Must be of type 'vector', not 'NULL'\n * checkmate::check_factor(data): Must be of type 'factor', not 'NULL'"),
@@ -515,7 +520,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_17050 <- xpectr::capture_side_effects(pair_extremes(data = c(1, 2, 3, 4), col = "A", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_17050 <- xpectr::capture_side_effects(pair_extremes(data = c(1, 2, 3, 4), col = "A", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_17050[['error']]),
xpectr::strip("1 assertions failed:\n * when 'data' is not a data.frame, 'col(s)' must be 'NULL'."),
@@ -529,7 +534,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data, factor_name
xpectr::set_test_seed(42)
# Assigning output
- output_14577 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "another_name", overwrite = TRUE)
+ output_14577 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "another_name", overwrite = TRUE)
# Testing class
expect_equal(
class(output_14577),
@@ -572,7 +577,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: col = "C"
xpectr::set_test_seed(42)
# Assigning output
- output_17191 <- pair_extremes(data = df, col = "C", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_17191 <- pair_extremes(data = df, col = "C", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_17191),
@@ -625,7 +630,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: col = "A"
xpectr::set_test_seed(42)
# Assigning output
- output_19346 <- pair_extremes(data = df, col = "A", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19346 <- pair_extremes(data = df, col = "A", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19346),
@@ -678,7 +683,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: col = "B"
xpectr::set_test_seed(42)
# Assigning output
- output_12554 <- pair_extremes(data = df, col = "B", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_12554 <- pair_extremes(data = df, col = "B", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_12554),
@@ -731,7 +736,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: unequal_method = "first"
xpectr::set_test_seed(42)
# Assigning output
- output_14622 <- pair_extremes(data = df, col = NULL, unequal_method = "first", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_14622 <- pair_extremes(data = df, col = NULL, unequal_method = "first", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_14622),
@@ -784,7 +789,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: unequal_method = "last"
xpectr::set_test_seed(42)
# Assigning output
- output_19400 <- pair_extremes(data = df, col = NULL, unequal_method = "last", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19400 <- pair_extremes(data = df, col = NULL, unequal_method = "last", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19400),
@@ -834,70 +839,410 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
fixed = TRUE)
# Testing pair_extremes(data = df, col = NULL, unequal...
- # Changed from baseline: unequal_method = NULL
+ # Changed from baseline: unequal_method = NA
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_19782 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = NULL, shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_19782 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = NA, num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_19782[['error']]),
- xpectr::strip("1 assertions failed:\n * Variable 'unequal_method': Must be of type 'string', not 'NULL'."),
+ xpectr::strip("1 assertions failed:\n * Variable 'unequal_method': May not be NA."),
fixed = TRUE)
expect_equal(
xpectr::strip(side_effects_19782[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: unequal_method = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_11174 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = NULL, num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_11174[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'unequal_method': Must be of type 'string', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_11174[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings = 2
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_14749 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_14749),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_14749[["index"]],
+ c(5, 4, 6, 2, 8, 1, 9, 3, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_14749[["A"]],
+ c(4, 5, 7, 8, 1, 9, 2, 3, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_14749[["B"]],
+ c(0.25543, 0.93467, 0.46229, 0.45774, 0.97823, 0.70506, 0.11749,
+ 0.71911, 0.94001),
+ tolerance = 1e-4)
+ expect_equal(
+ output_14749[["C"]],
+ c("E", "D", "F", "B", "H", "A", "I", "C", "G"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_14749),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_14749),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_14749),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_14749),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_14749)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_15603 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = NA, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15603[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'num_pairings': May not be NA."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15603[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_19040 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = NULL, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19040[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'num_pairings': Must be of type 'count', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19040[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings, balance
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_11387 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "spread", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_11387),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_11387[["index"]],
+ c(1, 9, 5, 3, 7, 2, 8, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11387[["A"]],
+ c(9, 2, 4, 3, 6, 8, 1, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11387[["B"]],
+ c(0.70506, 0.11749, 0.25543, 0.71911, 0.94001, 0.45774, 0.97823,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11387[["C"]],
+ c("A", "I", "E", "C", "G", "B", "H", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_11387),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_11387),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_11387),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_11387),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_11387)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings, balance
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19888 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "max", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_19888),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19888[["index"]],
+ c(1, 9, 5, 3, 7, 2, 8, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19888[["A"]],
+ c(9, 2, 4, 3, 6, 8, 1, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19888[["B"]],
+ c(0.70506, 0.11749, 0.25543, 0.71911, 0.94001, 0.45774, 0.97823,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19888[["C"]],
+ c("A", "I", "E", "C", "G", "B", "H", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_19888),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19888),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19888),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19888),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19888)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings, balance
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19466 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "min", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_19466),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19466[["index"]],
+ c(1, 9, 5, 3, 7, 2, 8, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19466[["A"]],
+ c(9, 2, 4, 3, 6, 8, 1, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19466[["B"]],
+ c(0.70506, 0.11749, 0.25543, 0.71911, 0.94001, 0.45774, 0.97823,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19466[["C"]],
+ c("A", "I", "E", "C", "G", "B", "H", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_19466),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19466),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19466),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19466),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19466)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: balance = "spread"
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_10824 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "spread", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_10824),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_10824[["index"]],
+ c(1, 9, 2, 8, 5, 3, 7, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10824[["A"]],
+ c(9, 2, 8, 1, 4, 3, 6, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10824[["B"]],
+ c(0.70506, 0.11749, 0.45774, 0.97823, 0.25543, 0.71911, 0.94001,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10824[["C"]],
+ c("A", "I", "B", "H", "E", "C", "G", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_10824),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_10824),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_10824),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_10824),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_10824)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: balance = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_15142 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = NA, shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15142[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'balance': Contains missing values (element 1)."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15142[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: balance = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_13902 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = NULL, shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_13902[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'balance': Must be of type 'character', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_13902[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
# Testing pair_extremes(data = df, col = NULL, unequal...
# Changed from baseline: shuffle_members = TRUE
xpectr::set_test_seed(42)
# Assigning output
- output_11174 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = TRUE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19057 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = TRUE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
- class(output_11174),
+ class(output_19057),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_11174[["index"]],
- c(9, 1, 8, 2, 5, 3, 7, 6, 4),
+ output_19057[["index"]],
+ c(6, 4, 2, 5, 8, 7, 3, 1, 9),
tolerance = 1e-4)
expect_equal(
- output_11174[["A"]],
- c(2, 9, 1, 8, 4, 3, 6, 7, 5),
+ output_19057[["A"]],
+ c(7, 5, 8, 4, 1, 6, 3, 9, 2),
tolerance = 1e-4)
expect_equal(
- output_11174[["B"]],
- c(0.11749, 0.70506, 0.97823, 0.45774, 0.25543, 0.71911, 0.94001,
- 0.46229, 0.93467),
+ output_19057[["B"]],
+ c(0.46229, 0.93467, 0.45774, 0.25543, 0.97823, 0.94001, 0.71911,
+ 0.70506, 0.11749),
tolerance = 1e-4)
expect_equal(
- output_11174[["C"]],
- c("I", "A", "H", "B", "E", "C", "G", "F", "D"),
+ output_19057[["C"]],
+ c("F", "D", "B", "E", "H", "G", "C", "A", "I"),
fixed = TRUE)
# Testing column names
expect_equal(
- names(output_11174),
+ names(output_19057),
c("index", "A", "B", "C"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_11174),
+ xpectr::element_classes(output_19057),
c("integer", "integer", "numeric", "character"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_11174),
+ xpectr::element_types(output_19057),
c("integer", "integer", "double", "character"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_11174),
+ dim(output_19057),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_11174)),
+ colnames(dplyr::group_keys(output_19057)),
character(0),
fixed = TRUE)
@@ -906,13 +1251,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_14749 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = NULL, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_14469 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = NULL, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_14749[['error']]),
+ xpectr::strip(side_effects_14469[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'shuffle_members': Must be of type 'logical flag', not 'NULL'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_14749[['error_class']]),
+ xpectr::strip(side_effects_14469[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -920,52 +1265,52 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: shuffle_members, shuf...
xpectr::set_test_seed(42)
# Assigning output
- output_15603 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = TRUE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
+ output_18360 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = TRUE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
- class(output_15603),
+ class(output_18360),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_15603[["index"]],
- c(5, 6, 4, 9, 1, 2, 8, 3, 7),
+ output_18360[["index"]],
+ c(7, 3, 2, 8, 5, 4, 6, 9, 1),
tolerance = 1e-4)
expect_equal(
- output_15603[["A"]],
- c(4, 7, 5, 2, 9, 8, 1, 3, 6),
+ output_18360[["A"]],
+ c(6, 3, 8, 1, 4, 5, 7, 2, 9),
tolerance = 1e-4)
expect_equal(
- output_15603[["B"]],
- c(0.25543, 0.46229, 0.93467, 0.11749, 0.70506, 0.45774, 0.97823,
- 0.71911, 0.94001),
+ output_18360[["B"]],
+ c(0.94001, 0.71911, 0.45774, 0.97823, 0.25543, 0.93467, 0.46229,
+ 0.11749, 0.70506),
tolerance = 1e-4)
expect_equal(
- output_15603[["C"]],
- c("E", "F", "D", "I", "A", "B", "H", "C", "G"),
+ output_18360[["C"]],
+ c("G", "C", "B", "H", "E", "D", "F", "I", "A"),
fixed = TRUE)
# Testing column names
expect_equal(
- names(output_15603),
+ names(output_18360),
c("index", "A", "B", "C"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_15603),
+ xpectr::element_classes(output_18360),
c("integer", "integer", "numeric", "character"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_15603),
+ xpectr::element_types(output_18360),
c("integer", "integer", "double", "character"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_15603),
+ dim(output_18360),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_15603)),
+ colnames(dplyr::group_keys(output_18360)),
character(0),
fixed = TRUE)
@@ -973,52 +1318,52 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: shuffle_pairs = TRUE
xpectr::set_test_seed(42)
# Assigning output
- output_19040 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
+ output_17375 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
- class(output_19040),
+ class(output_17375),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_19040[["index"]],
- c(5, 4, 6, 1, 9, 2, 8, 3, 7),
+ output_17375[["index"]],
+ c(9, 1, 2, 8, 5, 7, 3, 4, 6),
tolerance = 1e-4)
expect_equal(
- output_19040[["A"]],
- c(4, 5, 7, 9, 2, 8, 1, 3, 6),
+ output_17375[["A"]],
+ c(2, 9, 8, 1, 4, 6, 3, 5, 7),
tolerance = 1e-4)
expect_equal(
- output_19040[["B"]],
- c(0.25543, 0.93467, 0.46229, 0.70506, 0.11749, 0.45774, 0.97823,
- 0.71911, 0.94001),
+ output_17375[["B"]],
+ c(0.11749, 0.70506, 0.45774, 0.97823, 0.25543, 0.94001, 0.71911,
+ 0.93467, 0.46229),
tolerance = 1e-4)
expect_equal(
- output_19040[["C"]],
- c("E", "D", "F", "A", "I", "B", "H", "C", "G"),
+ output_17375[["C"]],
+ c("I", "A", "B", "H", "E", "G", "C", "D", "F"),
fixed = TRUE)
# Testing column names
expect_equal(
- names(output_19040),
+ names(output_17375),
c("index", "A", "B", "C"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_19040),
+ xpectr::element_classes(output_17375),
c("integer", "integer", "numeric", "character"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_19040),
+ xpectr::element_types(output_17375),
c("integer", "integer", "double", "character"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_19040),
+ dim(output_17375),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_19040)),
+ colnames(dplyr::group_keys(output_17375)),
character(0),
fixed = TRUE)
@@ -1027,13 +1372,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_11387 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = NULL, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_18110 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = NULL, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_11387[['error']]),
+ xpectr::strip(side_effects_18110[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'shuffle_pairs': Must be of type 'logical flag', not 'NULL'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_11387[['error_class']]),
+ xpectr::strip(side_effects_18110[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1041,56 +1386,56 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: factor_name = ".pair"
xpectr::set_test_seed(42)
# Assigning output
- output_19888 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = ".pair", overwrite = TRUE)
+ output_13881 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = ".pair", overwrite = TRUE)
# Testing class
expect_equal(
- class(output_19888),
+ class(output_13881),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_19888[["index"]],
+ output_13881[["index"]],
c(1, 9, 2, 8, 5, 3, 7, 4, 6),
tolerance = 1e-4)
expect_equal(
- output_19888[["A"]],
+ output_13881[["A"]],
c(9, 2, 8, 1, 4, 3, 6, 5, 7),
tolerance = 1e-4)
expect_equal(
- output_19888[["B"]],
+ output_13881[["B"]],
c(0.70506, 0.11749, 0.45774, 0.97823, 0.25543, 0.71911, 0.94001,
0.93467, 0.46229),
tolerance = 1e-4)
expect_equal(
- output_19888[["C"]],
+ output_13881[["C"]],
c("A", "I", "B", "H", "E", "C", "G", "D", "F"),
fixed = TRUE)
expect_equal(
- output_19888[[".pair"]],
+ output_13881[[".pair"]],
structure(c(1L, 1L, 2L, 2L, 3L, 4L, 4L, 5L, 5L), .Label = c("1",
"2", "3", "4", "5"), class = "factor"))
# Testing column names
expect_equal(
- names(output_19888),
+ names(output_13881),
c("index", "A", "B", "C", ".pair"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_19888),
+ xpectr::element_classes(output_13881),
c("integer", "integer", "numeric", "character", "factor"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_19888),
+ xpectr::element_types(output_13881),
c("integer", "integer", "double", "character", "integer"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_19888),
+ dim(output_13881),
c(9L, 5L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_19888)),
+ colnames(dplyr::group_keys(output_13881)),
character(0),
fixed = TRUE)
@@ -1098,52 +1443,52 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: factor_name = "A"
xpectr::set_test_seed(42)
# Assigning output
- output_19466 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = TRUE)
+ output_16851 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = TRUE)
# Testing class
expect_equal(
- class(output_19466),
+ class(output_16851),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_19466[["index"]],
+ output_16851[["index"]],
c(1, 9, 2, 8, 5, 3, 7, 4, 6),
tolerance = 1e-4)
expect_equal(
- output_19466[["A"]],
- structure(c(1L, 1L, 2L, 2L, 3L, 4L, 4L, 5L, 5L), .Label = c("1",
- "2", "3", "4", "5"), class = "factor"))
- expect_equal(
- output_19466[["B"]],
+ output_16851[["B"]],
c(0.70506, 0.11749, 0.45774, 0.97823, 0.25543, 0.71911, 0.94001,
0.93467, 0.46229),
tolerance = 1e-4)
expect_equal(
- output_19466[["C"]],
+ output_16851[["C"]],
c("A", "I", "B", "H", "E", "C", "G", "D", "F"),
fixed = TRUE)
+ expect_equal(
+ output_16851[["A"]],
+ structure(c(1L, 1L, 2L, 2L, 3L, 4L, 4L, 5L, 5L), .Label = c("1",
+ "2", "3", "4", "5"), class = "factor"))
# Testing column names
expect_equal(
- names(output_19466),
- c("index", "A", "B", "C"),
+ names(output_16851),
+ c("index", "B", "C", "A"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_19466),
- c("integer", "factor", "numeric", "character"),
+ xpectr::element_classes(output_16851),
+ c("integer", "numeric", "character", "factor"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_19466),
- c("integer", "integer", "double", "character"),
+ xpectr::element_types(output_16851),
+ c("integer", "double", "character", "integer"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_19466),
+ dim(output_16851),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_19466)),
+ colnames(dplyr::group_keys(output_16851)),
character(0),
fixed = TRUE)
@@ -1152,13 +1497,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_10824 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = 1, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_10039 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = 1, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_10824[['error']]),
+ xpectr::strip(side_effects_10039[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'factor_name': Must be of type 'string' (or 'NULL'), not 'double'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_10824[['error_class']]),
+ xpectr::strip(side_effects_10039[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1167,13 +1512,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_15142 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NA, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_18329 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NA, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_15142[['error']]),
+ xpectr::strip(side_effects_18329[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'factor_name': May not be NA."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_15142[['error_class']]),
+ xpectr::strip(side_effects_18329[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1182,13 +1527,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_13902 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = FALSE), reset_seed = TRUE)
+ side_effects_10073 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = FALSE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_13902[['error']]),
- xpectr::strip("1 assertions failed:\n * The column 'A' already exists and 'overwrite' is disabled."),
+ xpectr::strip(side_effects_10073[['error']]),
+ xpectr::strip("Adding these dimensions would overwrite existing columns: A."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_13902[['error_class']]),
+ xpectr::strip(side_effects_10073[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1197,13 +1542,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_19057 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = NULL), reset_seed = TRUE)
+ side_effects_12076 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = NULL), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_19057[['error']]),
+ xpectr::strip(side_effects_12076[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'overwrite': Must be of type 'logical flag', not 'NULL'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_19057[['error_class']]),
+ xpectr::strip(side_effects_12076[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1233,8 +1578,7 @@ test_that("testing pair_extremes_vec()", {
# Testing values
expect_equal(
output_19148,
- c(0.64175, 0.83045, 0.28614, 0.93708, 0.91481, 0.5191, 0.65699,
- 0.73659, 0.13467),
+ c(0.91480, 0.5190959, 0.736588, 0.65699, 0.6417, 0.8304476, 0.134667, 0.2861395, 0.937075),
tolerance = 1e-4)
# Testing names
expect_equal(
@@ -1252,4 +1596,43 @@ test_that("testing pair_extremes_vec()", {
## Finished testing 'pair_extremes_vec(data = runif(9), unequal_m...' ####
+
+
+ ## Testing 'pair_extremes_vec(data = runif(9), unequal_m...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_16932 <- pair_extremes_vec(data = runif(9),
+ unequal_method = "first",
+ num_pairings = 2)
+ # Testing class
+ expect_equal(
+ class(output_16932),
+ "numeric",
+ fixed = TRUE)
+ # Testing type
+ expect_type(
+ output_16932,
+ type = "double")
+ # Testing values
+ expect_equal(
+ output_16932,
+ c(0.13467, 0.28614, 0.93708, 0.64175, 0.83045, 0.5191, 0.91481,
+ 0.65699, 0.73659),
+ tolerance = 1e-4)
+ # Testing names
+ expect_equal(
+ names(output_16932),
+ NULL,
+ fixed = TRUE)
+ # Testing length
+ expect_equal(
+ length(output_16932),
+ 9L)
+ # Testing sum of element lengths
+ expect_equal(
+ sum(xpectr::element_lengths(output_16932)),
+ 9L)
+ ## Finished testing 'pair_extremes_vec(data = runif(9), unequal_m...' ####
+
})
diff --git a/tests/testthat/test_shuffle_hierarchy.R b/tests/testthat/test_shuffle_hierarchy.R
new file mode 100644
index 0000000..50d36bb
--- /dev/null
+++ b/tests/testthat/test_shuffle_hierarchy.R
@@ -0,0 +1,593 @@
+library(rearrr)
+context("shuffle_hierarchy")
+
+
+test_that("shuffle_hierarchy()", {
+ xpectr::set_test_seed(42)
+
+ df <- data.frame(
+ 'a' = rep(1:2, each = 8),
+ 'b' = rep(1:4, each = 4),
+ 'c' = rep(1:8, each = 2),
+ 'd' = 1:16
+ )
+
+ ## Testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(98)
+ # Assigning output
+ output_19148 <- shuffle_hierarchy(df, group_cols = c('a', 'b', 'c', 'd'))
+ # Testing class
+ expect_equal(
+ class(output_19148),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19148[["a"]],
+ c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["b"]],
+ c(4L, 4L, 4L, 4L, 3L, 3L, 3L, 3L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["c"]],
+ c(8L, 8L, 7L, 7L, 6L, 6L, 5L, 5L, 1L, 1L, 2L, 2L, 4L, 4L, 3L, 3L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["d"]],
+ c(15L, 16L, 13L, 14L, 12L, 11L, 10L, 9L, 1L, 2L, 3L, 4L, 7L, 8L, 5L, 6L),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_19148),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19148),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19148)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+
+
+
+ ## Testing 'shuffle_hierarchy( data = df, group_cols = c...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(98)
+ # Assigning output
+ output_16988 <- shuffle_hierarchy(
+ data = df,
+ group_cols = c('a', 'b', 'c', 'd'),
+ cols_to_shuffle = c('a', 'c')
+ )
+ # Testing class
+ expect_equal(
+ class(output_16988),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_16988[["a"]],
+ c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_16988[["b"]],
+ c(3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_16988[["c"]],
+ c(5L, 5L, 6L, 6L, 7L, 7L, 8L, 8L, 1L, 1L, 2L, 2L, 3L, 3L, 4L, 4L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_16988[["d"]],
+ c(9L, 10L, 11L, 12L, 13L, 14L, 15L, 16L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_16988),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_16988),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_16988),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_16988),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_16988)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy( data = df, group_cols = c...' ####
+
+ ## Testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_10268 <- shuffle_hierarchy(df, group_cols = c('a', 'b'), leaf_has_groups = FALSE)
+ # Testing class
+ expect_equal(
+ class(output_10268),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_10268[["a"]],
+ c(2, 2, 2, 2, 2, 2, 2, 2, 1, 1, 1, 1, 1, 1, 1, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10268[["b"]],
+ c(4, 4, 3, 3, 4, 3, 4, 3, 2, 2, 1, 2, 1, 2, 1, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10268[["c"]],
+ c(7, 7, 6, 6, 8, 5, 8, 5, 4, 4, 1, 3, 2, 3, 2, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10268[["d"]],
+ c(14, 13, 11, 12, 15, 9, 16, 10, 8, 7, 2, 5, 3, 6, 4, 1),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_10268),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_10268),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_10268),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_10268),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_10268)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+
+ # If leaf col is not in cols_to_shuffle
+ # last column should be considered a group (because it is not the leaf)
+
+ ## Testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19782 <- shuffle_hierarchy(df, group_cols = c('a', 'b'), cols_to_shuffle = 'a', leaf_has_groups = FALSE)
+ # Testing class
+ expect_equal(
+ class(output_19782),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19782[["a"]],
+ c(2, 2, 2, 2, 2, 2, 2, 2, 1, 1, 1, 1, 1, 1, 1, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19782[["b"]],
+ c(3, 3, 3, 3, 4, 4, 4, 4, 1, 1, 1, 1, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19782[["c"]],
+ c(5, 5, 6, 6, 7, 7, 8, 8, 1, 1, 2, 2, 3, 3, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19782[["d"]],
+ c(9, 10, 11, 12, 13, 14, 15, 16, 1, 2, 3, 4, 5, 6, 7, 8),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_19782),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19782),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19782),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19782),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19782)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+
+})
+
+
+test_that("fuzz testing shuffle_hierarchy()", {
+ xpectr::set_test_seed(42)
+
+ df <- data.frame(
+ 'a' = rep(1:2, each = 8),
+ 'b' = rep(1:4, each = 4),
+ 'c' = rep(1:8, each = 2),
+ 'd' = 1:16
+ )
+
+ # Generate expectations for 'shuffle_hierarchy'
+ # Tip: comment out the gxs_function() call
+ # so it is easy to regenerate the tests
+ xpectr::set_test_seed(42)
+ # xpectr::gxs_function(
+ # fn = shuffle_hierarchy,
+ # args_values = list(
+ # "data" = list(df, c(1,2,3), NA),
+ # "group_cols" = list(c('a', 'b', 'c', 'd'), c(1, 2, 3, 4), NA),
+ # "cols_to_shuffle" = list(c('a', 'b', 'c', 'd'), c('b', 'c'), 2, NA),
+ # "leaf_has_groups" = list(TRUE, FALSE, NA)
+ # ),
+ # indentation = 2,
+ # copy_env = FALSE
+ # )
+
+ ## Testing 'shuffle_hierarchy' ####
+ ## Initially generated by xpectr
+ # Testing different combinations of argument values
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19148 <- shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_19148),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19148[["a"]],
+ c(1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["b"]],
+ c(1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["c"]],
+ c(2, 2, 1, 1, 3, 3, 4, 4, 6, 6, 5, 5, 8, 8, 7, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["d"]],
+ c(3, 4, 2, 1, 6, 5, 8, 7, 11, 12, 10, 9, 15, 16, 14, 13),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_19148),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19148),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19148)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = c(1, 2, 3), group_c...
+ # Changed from baseline: data = c(1, 2, 3)
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_19370 <- xpectr::capture_side_effects(shuffle_hierarchy(data = c(1, 2, 3), group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19370[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'data': Must be of type 'data.frame', not 'double'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19370[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = NA, group_cols = c(...
+ # Changed from baseline: data = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_12861 <- xpectr::capture_side_effects(shuffle_hierarchy(data = NA, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12861[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'data': Must be of type 'data.frame', not 'logical'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12861[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = NULL, group_cols = ...
+ # Changed from baseline: data = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_18304 <- xpectr::capture_side_effects(shuffle_hierarchy(data = NULL, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_18304[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'data': Must be of type 'data.frame', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_18304[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: group_cols = c(1, 2, ...
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_16417 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c(1, 2, 3, 4), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16417[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'group_cols': Must be of type 'character', not 'double'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16417[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = NA...
+ # Changed from baseline: group_cols = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_15190 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = NA, cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15190[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'group_cols': Contains missing values (element 1)."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15190[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = NU...
+ # Changed from baseline: group_cols = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_17365 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = NULL, cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17365[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'group_cols': Must be of type 'character', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17365[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = c("...
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_11346 <- shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("b", "c"), leaf_has_groups = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_11346),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_11346[["a"]],
+ c(1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11346[["b"]],
+ c(2, 2, 2, 2, 1, 1, 1, 1, 3, 3, 3, 3, 4, 4, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11346[["c"]],
+ c(3, 3, 4, 4, 2, 2, 1, 1, 6, 6, 5, 5, 8, 8, 7, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11346[["d"]],
+ c(5, 6, 7, 8, 3, 4, 1, 2, 11, 12, 9, 10, 15, 16, 13, 14),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_11346),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_11346),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_11346),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_11346),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_11346)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = 2
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_16569 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = 2, leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16569[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'cols_to_shuffle': Must be of type 'character', not 'double'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16569[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_17050 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = NA, leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17050[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'cols_to_shuffle': Contains missing values (element 1)."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17050[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_14577 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = NULL, leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_14577[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'cols_to_shuffle': Must be of type 'character', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_14577[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: leaf_has_groups = FALSE
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_17191 <- shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = FALSE)
+ # Testing class
+ expect_equal(
+ class(output_17191),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_17191[["a"]],
+ c(1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_17191[["b"]],
+ c(1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_17191[["c"]],
+ c(2, 2, 1, 1, 3, 3, 4, 4, 6, 6, 5, 5, 8, 8, 7, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_17191[["d"]],
+ c(3, 4, 2, 1, 6, 5, 8, 7, 11, 12, 10, 9, 15, 16, 14, 13),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_17191),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_17191),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_17191),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_17191),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_17191)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: leaf_has_groups = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_19346 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = NA), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19346[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'leaf_has_groups': May not be NA."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19346[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: leaf_has_groups = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_12554 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = NULL), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12554[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'leaf_has_groups': Must be of type 'logical flag', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12554[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ ## Finished testing 'shuffle_hierarchy' ####
+ #
+
+})
+
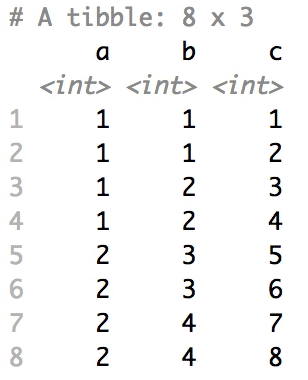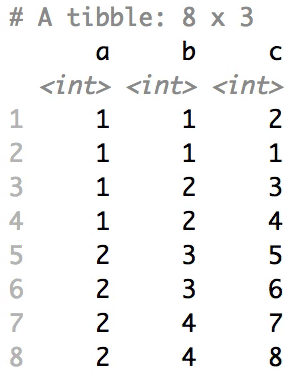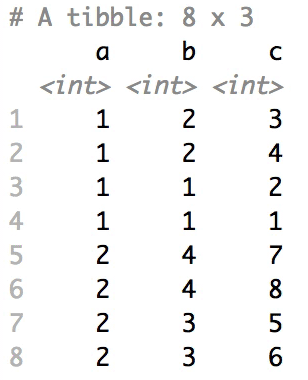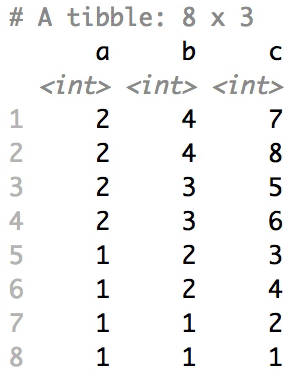 +
+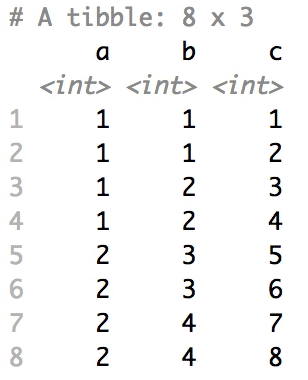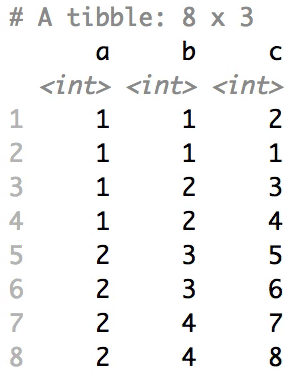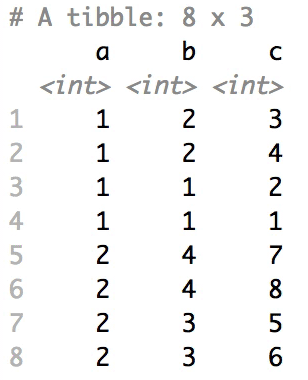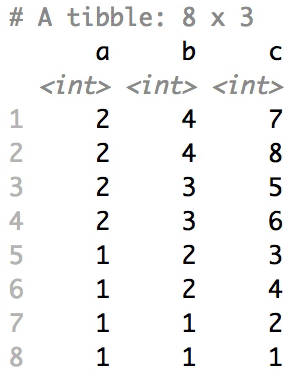 +
+ +### Shuffle Hierarchy
+
+When having a `data.frame` with multiple grouping columns, we can
+shuffle them one column (hierarchical level) at a time:
+
+``` r
+# Shuffle a given data frame 'df'
+shuffle_hierarchy(df, group_cols = c("a", "b", "c"))
+```
+
+The columns are shuffled one at a time, as
+so:
+
+
+### Shuffle Hierarchy
+
+When having a `data.frame` with multiple grouping columns, we can
+shuffle them one column (hierarchical level) at a time:
+
+``` r
+# Shuffle a given data frame 'df'
+shuffle_hierarchy(df, group_cols = c("a", "b", "c"))
+```
+
+The columns are shuffled one at a time, as
+so:
+
+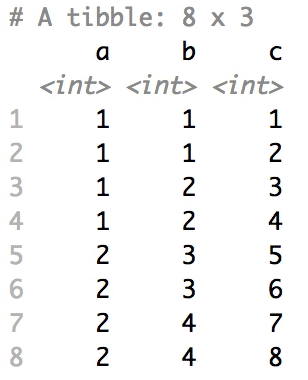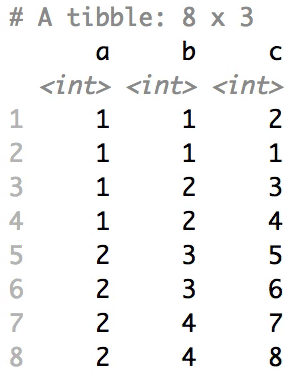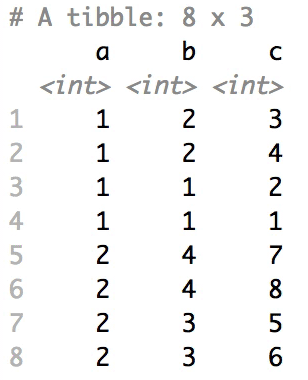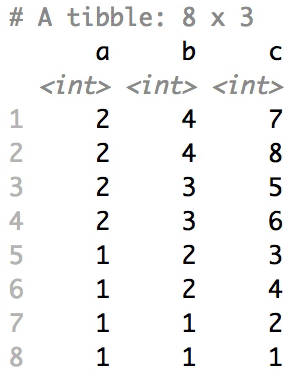 +
+
+
+ +
+ 3-dimensional rotation:
@@ -353,7 +374,7 @@ rotate_3d(
#> 12 12 18 0.505 -1.67 9.88 -5.04
3-dimensional rotation:
@@ -353,7 +374,7 @@ rotate_3d(
#> 12 12 18 0.505 -1.67 9.88 -5.04  ### Swirl values
@@ -461,7 +482,7 @@ expand_distances(
2d
expansion:
-
### Swirl values
@@ -461,7 +482,7 @@ expand_distances(
2d
expansion:
- +
+ Expand differently in each axis:
@@ -489,7 +510,7 @@ expand_distances_each(
#> 10 0.361 0.169 -0.466 0.251
Expand differently in each axis:
@@ -489,7 +510,7 @@ expand_distances_each(
#> 10 0.361 0.169 -0.466 0.251  +
+ ### Cluster groups
@@ -525,7 +546,7 @@ cluster_groups(
#> # … with 40 more rows
```
-
### Cluster groups
@@ -525,7 +546,7 @@ cluster_groups(
#> # … with 40 more rows
```
- +
+ ### Dim values
@@ -558,7 +579,7 @@ df_clustered %>%
#> # … with 40 more rows
```
-
### Dim values
@@ -558,7 +579,7 @@ df_clustered %>%
#> # … with 40 more rows
```
- +
+ ### Flip values
@@ -587,7 +608,7 @@ flip_values(
#> 10 0.0618 1.14
### Flip values
@@ -587,7 +608,7 @@ flip_values(
#> 10 0.0618 1.14  +
+ ## Forming examples
@@ -722,7 +743,7 @@ generate_clusters(
#> # … with 40 more rows
```
-
## Forming examples
@@ -722,7 +743,7 @@ generate_clusters(
#> # … with 40 more rows
```
- +
+ ## Utilities
diff --git a/cran-comments.md b/cran-comments.md
index 4b4fe6a..02c18c2 100644
--- a/cran-comments.md
+++ b/cran-comments.md
@@ -5,9 +5,4 @@
## R CMD check results
-0 errors | 0 warnings | 1 note
-
-* This is a new release.
-* Removed examples for unexported functions
-* Made examples testable in most cases
-* Replaced <<- assignment with specific assignment to the right environment
+0 errors | 0 warnings | 0 notes
diff --git a/man/center_max.Rd b/man/center_max.Rd
index 729ce93..0a316f0 100644
--- a/man/center_max.Rd
+++ b/man/center_max.Rd
@@ -84,7 +84,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/center_min.Rd b/man/center_min.Rd
index e965bf7..0131e96 100644
--- a/man/center_min.Rd
+++ b/man/center_min.Rd
@@ -84,7 +84,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/closest_to.Rd b/man/closest_to.Rd
index 3c8a1d1..a585d95 100644
--- a/man/closest_to.Rd
+++ b/man/closest_to.Rd
@@ -165,7 +165,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
Other distance functions:
\code{\link{dim_values}()},
diff --git a/man/extreme_pairing_rearranger_.Rd b/man/extreme_pairing_rearranger_.Rd
index cb83c7e..4ede22f 100644
--- a/man/extreme_pairing_rearranger_.Rd
+++ b/man/extreme_pairing_rearranger_.Rd
@@ -11,6 +11,7 @@ extreme_pairing_rearranger_(
shuffle_members = FALSE,
shuffle_pairs = FALSE,
num_pairings = 1,
+ balance = "mean",
factor_name = ".pair",
overwrite = FALSE
)
@@ -31,7 +32,7 @@ The first group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -50,7 +51,7 @@ The middle group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -68,7 +69,7 @@ The last group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -86,15 +87,40 @@ And are \strong{ordered as}:
\item{shuffle_pairs}{Whether to shuffle the pairs. (Logical)}
+\item{num_pairings}{Number of pairings to perform (recursively). At least \code{1}.
+
+Based on \code{`balance`}, the secondary pairings perform extreme pairing on either the
+\emph{sum}, \emph{absolute difference}, \emph{min}, or \emph{max} of the pair elements.}
+
+\item{balance}{What to balance pairs for in a given \emph{secondary} pairing.
+Either \code{"mean"}, \code{"spread"}, \code{"min"}, or \code{"max"}.
+Can be a single string used for all secondary pairings
+or one for each secondary pairing (\code{`num_pairings` - 1}).
+
+The first pairing always pairs the actual element values.
+
+\subsection{mean}{
+Pairs have similar means. The values in the pairs from the previous pairing
+are aggregated with \code{`sum()`} and paired.
+}
+\subsection{spread}{
+Pairs have similar spread (e.g. standard deviations). The values in the pairs from the previous pairing
+are aggregated with \code{`sum(abs(diff()))`} and paired.
+}
+\subsection{min / max}{
+Pairs have similar minimum / maximum values. The values in the pairs from the previous pairing
+are aggregated with \code{`min()`} / \code{`max()`} and paired.
+}}
+
\item{factor_name}{Name of new column with the sorting factor. If \code{NULL}, no column is added.}
\item{overwrite}{Whether to allow overwriting of existing columns. (Logical)}
}
\value{
The sorted \code{data.frame} (\code{tibble}) / \code{vector}.
-Optionally with the sorting factor added.
+Optionally with the sorting factor(s) added.
-When \code{`data`} is a \code{vector} and \code{`keep_factors`} is \code{FALSE},
+When \code{`data`} is a \code{vector} and \code{`factor_name`} is \code{NULL},
the output will be a \code{vector}. Otherwise, a \code{data.frame}.
}
\description{
diff --git a/man/figures/README-unnamed-chunk-19-1.png b/man/figures/README-unnamed-chunk-19-1.png
new file mode 100644
index 0000000..e6e93ee
Binary files /dev/null and b/man/figures/README-unnamed-chunk-19-1.png differ
diff --git a/man/figures/README-unnamed-chunk-21-1.png b/man/figures/README-unnamed-chunk-21-1.png
new file mode 100644
index 0000000..766574a
Binary files /dev/null and b/man/figures/README-unnamed-chunk-21-1.png differ
diff --git a/man/figures/README-unnamed-chunk-27-1.png b/man/figures/README-unnamed-chunk-27-1.png
new file mode 100644
index 0000000..927a9f9
Binary files /dev/null and b/man/figures/README-unnamed-chunk-27-1.png differ
diff --git a/man/figures/README-unnamed-chunk-29-1.png b/man/figures/README-unnamed-chunk-29-1.png
new file mode 100644
index 0000000..3c5a2ff
Binary files /dev/null and b/man/figures/README-unnamed-chunk-29-1.png differ
diff --git a/man/figures/README-unnamed-chunk-31-1.png b/man/figures/README-unnamed-chunk-31-1.png
new file mode 100644
index 0000000..5e97abd
Binary files /dev/null and b/man/figures/README-unnamed-chunk-31-1.png differ
diff --git a/man/figures/README-unnamed-chunk-33-1.png b/man/figures/README-unnamed-chunk-33-1.png
new file mode 100644
index 0000000..2487964
Binary files /dev/null and b/man/figures/README-unnamed-chunk-33-1.png differ
diff --git a/man/figures/README-unnamed-chunk-35-1.png b/man/figures/README-unnamed-chunk-35-1.png
new file mode 100644
index 0000000..0e4b3d0
Binary files /dev/null and b/man/figures/README-unnamed-chunk-35-1.png differ
diff --git a/man/figures/README-unnamed-chunk-46-1.png b/man/figures/README-unnamed-chunk-46-1.png
new file mode 100644
index 0000000..24b9de6
Binary files /dev/null and b/man/figures/README-unnamed-chunk-46-1.png differ
diff --git a/man/furthest_from.Rd b/man/furthest_from.Rd
index 4f7980a..17afb31 100644
--- a/man/furthest_from.Rd
+++ b/man/furthest_from.Rd
@@ -166,7 +166,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
Other distance functions:
\code{\link{closest_to}()},
diff --git a/man/pair_extremes.Rd b/man/pair_extremes.Rd
index b0eb76b..08fe718 100644
--- a/man/pair_extremes.Rd
+++ b/man/pair_extremes.Rd
@@ -9,15 +9,19 @@ pair_extremes(
data,
col = NULL,
unequal_method = "middle",
+ num_pairings = 1,
+ balance = "mean",
shuffle_members = FALSE,
shuffle_pairs = FALSE,
- factor_name = ".pair",
+ factor_name = ifelse(num_pairings == 1, ".pair", ".pairing"),
overwrite = FALSE
)
pair_extremes_vec(
data,
unequal_method = "middle",
+ num_pairings = 1,
+ balance = "mean",
shuffle_members = FALSE,
shuffle_pairs = FALSE
)
@@ -38,7 +42,7 @@ The first group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -57,7 +61,7 @@ The middle group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -75,7 +79,7 @@ The last group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -89,6 +93,31 @@ And are \strong{ordered as}:
}}
+\item{num_pairings}{Number of pairings to perform (recursively). At least \code{1}.
+
+Based on \code{`balance`}, the secondary pairings perform extreme pairing on either the
+\emph{sum}, \emph{absolute difference}, \emph{min}, or \emph{max} of the pair elements.}
+
+\item{balance}{What to balance pairs for in a given \emph{secondary} pairing.
+Either \code{"mean"}, \code{"spread"}, \code{"min"}, or \code{"max"}.
+Can be a single string used for all secondary pairings
+or one for each secondary pairing (\code{`num_pairings` - 1}).
+
+The first pairing always pairs the actual element values.
+
+\subsection{mean}{
+Pairs have similar means. The values in the pairs from the previous pairing
+are aggregated with \code{`sum()`} and paired.
+}
+\subsection{spread}{
+Pairs have similar spread (e.g. standard deviations). The values in the pairs from the previous pairing
+are aggregated with \code{`sum(abs(diff()))`} and paired.
+}
+\subsection{min / max}{
+Pairs have similar minimum / maximum values. The values in the pairs from the previous pairing
+are aggregated with \code{`min()`} / \code{`max()`} and paired.
+}}
+
\item{shuffle_members}{Whether to shuffle the pair members. (Logical)}
\item{shuffle_pairs}{Whether to shuffle the pairs. (Logical)}
@@ -166,6 +195,9 @@ pair_extremes(df, col = "A", shuffle_members = TRUE)
# Shuffle the order of the pairs
pair_extremes(df, col = "A", shuffle_pairs = TRUE)
+# Use recursive pairing
+pair_extremes(df, col = "A", num_pairings = 2)
+
# Grouped by G
df \%>\%
dplyr::select(G, A) \%>\% # For clarity
@@ -197,7 +229,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/position_max.Rd b/man/position_max.Rd
index 751e6ea..06a8d0c 100644
--- a/man/position_max.Rd
+++ b/man/position_max.Rd
@@ -89,7 +89,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/position_min.Rd b/man/position_min.Rd
index f1e85ae..e12c003 100644
--- a/man/position_min.Rd
+++ b/man/position_min.Rd
@@ -91,7 +91,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_max}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/rev_windows.Rd b/man/rev_windows.Rd
index a81f40b..59d46ae 100644
--- a/man/rev_windows.Rd
+++ b/man/rev_windows.Rd
@@ -91,7 +91,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_max}()},
\code{\link{position_min}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/roll_elements.Rd b/man/roll_elements.Rd
index f4c6bd8..e2ef28f 100644
--- a/man/roll_elements.Rd
+++ b/man/roll_elements.Rd
@@ -137,7 +137,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_max}()},
\code{\link{position_min}()},
-\code{\link{rev_windows}()}
+\code{\link{rev_windows}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/shuffle_hierarchy.Rd b/man/shuffle_hierarchy.Rd
new file mode 100644
index 0000000..dd49c84
--- /dev/null
+++ b/man/shuffle_hierarchy.Rd
@@ -0,0 +1,84 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/shuffle_hierarchy.R
+\name{shuffle_hierarchy}
+\alias{shuffle_hierarchy}
+\title{Shuffle multi-column hierarchy of groups}
+\usage{
+shuffle_hierarchy(
+ data,
+ group_cols,
+ cols_to_shuffle = group_cols,
+ leaf_has_groups = TRUE
+)
+}
+\arguments{
+\item{data}{\code{data.frame}.}
+
+\item{group_cols}{Names of columns making up the group hierarchy.
+The last column is the \emph{leaf} and is shuffled first (if also in \code{`cols_to_shuffle`}).}
+
+\item{cols_to_shuffle}{Names of columns to shuffle hierarchically.
+By default, all the \code{`group_cols`} are shuffled.}
+
+\item{leaf_has_groups}{Whether the leaf column contains groups or values. (Logical)
+
+When the elements are \emph{group identifiers}, they are ordered sequentially and shuffled together.
+
+When the elements are \emph{values}, they are simply shuffled.}
+}
+\value{
+The shuffled \code{data.frame} (\code{tibble}).
+}
+\description{
+\Sexpr[results=rd, stage=render]{lifecycle::badge("experimental")}
+
+Shuffles a tree/hierarchy of groups, one column at a time.
+The levels in the last ("leaf") column are shuffled first, then the second-last column, and so on.
+Elements of the same group are ordered sequentially.
+}
+\examples{
+# Attach packages
+library(rearrr)
+library(dplyr)
+
+df <- data.frame(
+ 'a' = rep(1:4, each = 4),
+ 'b' = rep(1:8, each = 2),
+ 'c' = 1:16
+)
+
+# Set seed for reproducibility
+set.seed(2)
+
+# Shuffle all columns
+shuffle_hierarchy(df, group_cols = c('a', 'b', 'c'))
+
+# Don't shuffle 'b' but keep grouping by it
+# So 'c' will be shuffled within each group in 'b'
+shuffle_hierarchy(
+ data = df,
+ group_cols = c('a', 'b', 'c'),
+ cols_to_shuffle = c('a', 'c')
+)
+
+# Shuffle 'b' as if it's not a group column
+# so elements are independent within their group
+# (i.e. same-valued elements are not necessarily ordered sequentially)
+shuffle_hierarchy(df, group_cols = c('a', 'b'), leaf_has_groups = FALSE)
+}
+\seealso{
+Other rearrange functions:
+\code{\link{center_max}()},
+\code{\link{center_min}()},
+\code{\link{closest_to}()},
+\code{\link{furthest_from}()},
+\code{\link{pair_extremes}()},
+\code{\link{position_max}()},
+\code{\link{position_min}()},
+\code{\link{rev_windows}()},
+\code{\link{roll_elements}()}
+}
+\author{
+Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
+}
+\concept{rearrange functions}
diff --git a/tests/testthat/test_pair_extremes.R b/tests/testthat/test_pair_extremes.R
index dda4aa2..52c915f 100644
--- a/tests/testthat/test_pair_extremes.R
+++ b/tests/testthat/test_pair_extremes.R
@@ -2,7 +2,7 @@ library(rearrr)
context("pair_extremes()")
-test_that("rearrange() with method pair_extremes works", {
+test_that("pair_extremes() works", {
# Create data frame
xpectr::set_test_seed(1)
@@ -70,7 +70,7 @@ test_that("rearrange() with method pair_extremes works", {
expect_equal(df_rearranged$score, c(69, 140, 79, 92, 85, 87))
})
-test_that("rearrange() with method pair_extremes throws expected errors", {
+test_that("pair_extremes() throws expected errors", {
df <- data.frame("a" = c(1,2,3))
## Testing 'rearrange(df, method = "pair_extremes", uneq...' ####
@@ -209,7 +209,9 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# "data" = list(df, head(df, 8), c(1,2,3,4,5,6,7), c(1,2,3,4),
# factor(c(1,2,3,4,5,6,7,1,2,3)), list(1,2,3), NA, 1),
# "col" = list(NULL, "A", "B", "C"),
- # "unequal_method" = list("middle", "first", "last"),
+ # "unequal_method" = list("middle", "first", "last", NA),
+ # "num_pairings" = list(1, 2, NA),
+ # "balance" = list("mean", "spread", NA),
# "shuffle_members" = list(FALSE, TRUE),
# "shuffle_pairs" = list(FALSE, TRUE),
# "factor_name" = list(NULL, ".pair", "A", 1, NA),
@@ -219,7 +221,10 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# list("data" = c(1,2,3,4), "col" = "A"),
# list("data" = c(1,2,3,4), "factor_name" = "another_name"),
# list("shuffle_members" = TRUE, "shuffle_pairs" = TRUE),
- # list("factor_name" = "A", "overwrite" = FALSE)
+ # list("factor_name" = "A", "overwrite" = FALSE),
+ # list("num_pairings" = 2, "balance" = "spread"),
+ # list("num_pairings" = 2, "balance" = "max"),
+ # list("num_pairings" = 2, "balance" = "min")
# ),
# indentation = 2
# )
@@ -232,7 +237,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Testing pair_extremes(data = df, col = NULL, unequal...
xpectr::set_test_seed(42)
# Assigning output
- output_19148 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19148 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19148),
@@ -285,7 +290,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = head(df, 8)
xpectr::set_test_seed(42)
# Assigning output
- output_19370 <- pair_extremes(data = head(df, 8), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19370 <- pair_extremes(data = head(df, 8), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19370),
@@ -338,7 +343,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = c(1, 2, 3, 4, ...
xpectr::set_test_seed(42)
# Assigning output
- output_12861 <- pair_extremes(data = c(1, 2, 3, 4, 5, 6, 7), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_12861 <- pair_extremes(data = c(1, 2, 3, 4, 5, 6, 7), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_12861),
@@ -371,7 +376,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = c(1, 2, 3, 4)
xpectr::set_test_seed(42)
# Assigning output
- output_18304 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_18304 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_18304),
@@ -404,7 +409,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = factor(c(1, 2,...
xpectr::set_test_seed(42)
# Assigning output
- output_16417 <- pair_extremes(data = factor(c(1, 2, 3, 4, 5, 6, 7, 1, 2, 3)), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_16417 <- pair_extremes(data = factor(c(1, 2, 3, 4, 5, 6, 7, 1, 2, 3)), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing is factor
expect_true(
is.factor(output_16417))
@@ -437,7 +442,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_15190 <- xpectr::capture_side_effects(pair_extremes(data = list(1, 2, 3), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_15190 <- xpectr::capture_side_effects(pair_extremes(data = list(1, 2, 3), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_15190[['error']]),
xpectr::strip("1 assertions failed:\n * when 'data' is not a data.frame, it cannot be a list."),
@@ -452,7 +457,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_17365 <- xpectr::capture_side_effects(pair_extremes(data = NA, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_17365 <- xpectr::capture_side_effects(pair_extremes(data = NA, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_17365[['error']]),
xpectr::strip("Assertion failed. One of the following must apply:\n * checkmate::check_data_frame(data): Must be of type 'data.frame', not 'logical'\n * checkmate::check_vector(data): Contains missing values (element 1)\n * checkmate::check_factor(data): Contains missing values (element 1)"),
@@ -466,7 +471,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = 1
xpectr::set_test_seed(42)
# Assigning output
- output_11346 <- pair_extremes(data = 1, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_11346 <- pair_extremes(data = 1, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_11346),
@@ -500,7 +505,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_16569 <- xpectr::capture_side_effects(pair_extremes(data = NULL, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_16569 <- xpectr::capture_side_effects(pair_extremes(data = NULL, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_16569[['error']]),
xpectr::strip("Assertion failed. One of the following must apply:\n * checkmate::check_data_frame(data): Must be of type 'data.frame', not 'NULL'\n * checkmate::check_vector(data): Must be of type 'vector', not 'NULL'\n * checkmate::check_factor(data): Must be of type 'factor', not 'NULL'"),
@@ -515,7 +520,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_17050 <- xpectr::capture_side_effects(pair_extremes(data = c(1, 2, 3, 4), col = "A", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_17050 <- xpectr::capture_side_effects(pair_extremes(data = c(1, 2, 3, 4), col = "A", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_17050[['error']]),
xpectr::strip("1 assertions failed:\n * when 'data' is not a data.frame, 'col(s)' must be 'NULL'."),
@@ -529,7 +534,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data, factor_name
xpectr::set_test_seed(42)
# Assigning output
- output_14577 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "another_name", overwrite = TRUE)
+ output_14577 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "another_name", overwrite = TRUE)
# Testing class
expect_equal(
class(output_14577),
@@ -572,7 +577,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: col = "C"
xpectr::set_test_seed(42)
# Assigning output
- output_17191 <- pair_extremes(data = df, col = "C", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_17191 <- pair_extremes(data = df, col = "C", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_17191),
@@ -625,7 +630,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: col = "A"
xpectr::set_test_seed(42)
# Assigning output
- output_19346 <- pair_extremes(data = df, col = "A", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19346 <- pair_extremes(data = df, col = "A", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19346),
@@ -678,7 +683,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: col = "B"
xpectr::set_test_seed(42)
# Assigning output
- output_12554 <- pair_extremes(data = df, col = "B", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_12554 <- pair_extremes(data = df, col = "B", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_12554),
@@ -731,7 +736,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: unequal_method = "first"
xpectr::set_test_seed(42)
# Assigning output
- output_14622 <- pair_extremes(data = df, col = NULL, unequal_method = "first", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_14622 <- pair_extremes(data = df, col = NULL, unequal_method = "first", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_14622),
@@ -784,7 +789,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: unequal_method = "last"
xpectr::set_test_seed(42)
# Assigning output
- output_19400 <- pair_extremes(data = df, col = NULL, unequal_method = "last", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19400 <- pair_extremes(data = df, col = NULL, unequal_method = "last", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19400),
@@ -834,70 +839,410 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
fixed = TRUE)
# Testing pair_extremes(data = df, col = NULL, unequal...
- # Changed from baseline: unequal_method = NULL
+ # Changed from baseline: unequal_method = NA
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_19782 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = NULL, shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_19782 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = NA, num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_19782[['error']]),
- xpectr::strip("1 assertions failed:\n * Variable 'unequal_method': Must be of type 'string', not 'NULL'."),
+ xpectr::strip("1 assertions failed:\n * Variable 'unequal_method': May not be NA."),
fixed = TRUE)
expect_equal(
xpectr::strip(side_effects_19782[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: unequal_method = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_11174 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = NULL, num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_11174[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'unequal_method': Must be of type 'string', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_11174[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings = 2
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_14749 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_14749),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_14749[["index"]],
+ c(5, 4, 6, 2, 8, 1, 9, 3, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_14749[["A"]],
+ c(4, 5, 7, 8, 1, 9, 2, 3, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_14749[["B"]],
+ c(0.25543, 0.93467, 0.46229, 0.45774, 0.97823, 0.70506, 0.11749,
+ 0.71911, 0.94001),
+ tolerance = 1e-4)
+ expect_equal(
+ output_14749[["C"]],
+ c("E", "D", "F", "B", "H", "A", "I", "C", "G"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_14749),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_14749),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_14749),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_14749),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_14749)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_15603 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = NA, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15603[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'num_pairings': May not be NA."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15603[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_19040 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = NULL, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19040[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'num_pairings': Must be of type 'count', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19040[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings, balance
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_11387 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "spread", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_11387),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_11387[["index"]],
+ c(1, 9, 5, 3, 7, 2, 8, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11387[["A"]],
+ c(9, 2, 4, 3, 6, 8, 1, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11387[["B"]],
+ c(0.70506, 0.11749, 0.25543, 0.71911, 0.94001, 0.45774, 0.97823,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11387[["C"]],
+ c("A", "I", "E", "C", "G", "B", "H", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_11387),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_11387),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_11387),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_11387),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_11387)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings, balance
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19888 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "max", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_19888),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19888[["index"]],
+ c(1, 9, 5, 3, 7, 2, 8, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19888[["A"]],
+ c(9, 2, 4, 3, 6, 8, 1, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19888[["B"]],
+ c(0.70506, 0.11749, 0.25543, 0.71911, 0.94001, 0.45774, 0.97823,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19888[["C"]],
+ c("A", "I", "E", "C", "G", "B", "H", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_19888),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19888),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19888),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19888),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19888)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings, balance
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19466 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "min", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_19466),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19466[["index"]],
+ c(1, 9, 5, 3, 7, 2, 8, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19466[["A"]],
+ c(9, 2, 4, 3, 6, 8, 1, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19466[["B"]],
+ c(0.70506, 0.11749, 0.25543, 0.71911, 0.94001, 0.45774, 0.97823,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19466[["C"]],
+ c("A", "I", "E", "C", "G", "B", "H", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_19466),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19466),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19466),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19466),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19466)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: balance = "spread"
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_10824 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "spread", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_10824),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_10824[["index"]],
+ c(1, 9, 2, 8, 5, 3, 7, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10824[["A"]],
+ c(9, 2, 8, 1, 4, 3, 6, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10824[["B"]],
+ c(0.70506, 0.11749, 0.45774, 0.97823, 0.25543, 0.71911, 0.94001,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10824[["C"]],
+ c("A", "I", "B", "H", "E", "C", "G", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_10824),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_10824),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_10824),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_10824),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_10824)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: balance = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_15142 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = NA, shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15142[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'balance': Contains missing values (element 1)."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15142[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: balance = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_13902 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = NULL, shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_13902[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'balance': Must be of type 'character', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_13902[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
# Testing pair_extremes(data = df, col = NULL, unequal...
# Changed from baseline: shuffle_members = TRUE
xpectr::set_test_seed(42)
# Assigning output
- output_11174 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = TRUE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19057 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = TRUE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
- class(output_11174),
+ class(output_19057),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_11174[["index"]],
- c(9, 1, 8, 2, 5, 3, 7, 6, 4),
+ output_19057[["index"]],
+ c(6, 4, 2, 5, 8, 7, 3, 1, 9),
tolerance = 1e-4)
expect_equal(
- output_11174[["A"]],
- c(2, 9, 1, 8, 4, 3, 6, 7, 5),
+ output_19057[["A"]],
+ c(7, 5, 8, 4, 1, 6, 3, 9, 2),
tolerance = 1e-4)
expect_equal(
- output_11174[["B"]],
- c(0.11749, 0.70506, 0.97823, 0.45774, 0.25543, 0.71911, 0.94001,
- 0.46229, 0.93467),
+ output_19057[["B"]],
+ c(0.46229, 0.93467, 0.45774, 0.25543, 0.97823, 0.94001, 0.71911,
+ 0.70506, 0.11749),
tolerance = 1e-4)
expect_equal(
- output_11174[["C"]],
- c("I", "A", "H", "B", "E", "C", "G", "F", "D"),
+ output_19057[["C"]],
+ c("F", "D", "B", "E", "H", "G", "C", "A", "I"),
fixed = TRUE)
# Testing column names
expect_equal(
- names(output_11174),
+ names(output_19057),
c("index", "A", "B", "C"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_11174),
+ xpectr::element_classes(output_19057),
c("integer", "integer", "numeric", "character"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_11174),
+ xpectr::element_types(output_19057),
c("integer", "integer", "double", "character"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_11174),
+ dim(output_19057),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_11174)),
+ colnames(dplyr::group_keys(output_19057)),
character(0),
fixed = TRUE)
@@ -906,13 +1251,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_14749 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = NULL, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_14469 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = NULL, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_14749[['error']]),
+ xpectr::strip(side_effects_14469[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'shuffle_members': Must be of type 'logical flag', not 'NULL'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_14749[['error_class']]),
+ xpectr::strip(side_effects_14469[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -920,52 +1265,52 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: shuffle_members, shuf...
xpectr::set_test_seed(42)
# Assigning output
- output_15603 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = TRUE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
+ output_18360 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = TRUE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
- class(output_15603),
+ class(output_18360),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_15603[["index"]],
- c(5, 6, 4, 9, 1, 2, 8, 3, 7),
+ output_18360[["index"]],
+ c(7, 3, 2, 8, 5, 4, 6, 9, 1),
tolerance = 1e-4)
expect_equal(
- output_15603[["A"]],
- c(4, 7, 5, 2, 9, 8, 1, 3, 6),
+ output_18360[["A"]],
+ c(6, 3, 8, 1, 4, 5, 7, 2, 9),
tolerance = 1e-4)
expect_equal(
- output_15603[["B"]],
- c(0.25543, 0.46229, 0.93467, 0.11749, 0.70506, 0.45774, 0.97823,
- 0.71911, 0.94001),
+ output_18360[["B"]],
+ c(0.94001, 0.71911, 0.45774, 0.97823, 0.25543, 0.93467, 0.46229,
+ 0.11749, 0.70506),
tolerance = 1e-4)
expect_equal(
- output_15603[["C"]],
- c("E", "F", "D", "I", "A", "B", "H", "C", "G"),
+ output_18360[["C"]],
+ c("G", "C", "B", "H", "E", "D", "F", "I", "A"),
fixed = TRUE)
# Testing column names
expect_equal(
- names(output_15603),
+ names(output_18360),
c("index", "A", "B", "C"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_15603),
+ xpectr::element_classes(output_18360),
c("integer", "integer", "numeric", "character"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_15603),
+ xpectr::element_types(output_18360),
c("integer", "integer", "double", "character"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_15603),
+ dim(output_18360),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_15603)),
+ colnames(dplyr::group_keys(output_18360)),
character(0),
fixed = TRUE)
@@ -973,52 +1318,52 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: shuffle_pairs = TRUE
xpectr::set_test_seed(42)
# Assigning output
- output_19040 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
+ output_17375 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
- class(output_19040),
+ class(output_17375),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_19040[["index"]],
- c(5, 4, 6, 1, 9, 2, 8, 3, 7),
+ output_17375[["index"]],
+ c(9, 1, 2, 8, 5, 7, 3, 4, 6),
tolerance = 1e-4)
expect_equal(
- output_19040[["A"]],
- c(4, 5, 7, 9, 2, 8, 1, 3, 6),
+ output_17375[["A"]],
+ c(2, 9, 8, 1, 4, 6, 3, 5, 7),
tolerance = 1e-4)
expect_equal(
- output_19040[["B"]],
- c(0.25543, 0.93467, 0.46229, 0.70506, 0.11749, 0.45774, 0.97823,
- 0.71911, 0.94001),
+ output_17375[["B"]],
+ c(0.11749, 0.70506, 0.45774, 0.97823, 0.25543, 0.94001, 0.71911,
+ 0.93467, 0.46229),
tolerance = 1e-4)
expect_equal(
- output_19040[["C"]],
- c("E", "D", "F", "A", "I", "B", "H", "C", "G"),
+ output_17375[["C"]],
+ c("I", "A", "B", "H", "E", "G", "C", "D", "F"),
fixed = TRUE)
# Testing column names
expect_equal(
- names(output_19040),
+ names(output_17375),
c("index", "A", "B", "C"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_19040),
+ xpectr::element_classes(output_17375),
c("integer", "integer", "numeric", "character"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_19040),
+ xpectr::element_types(output_17375),
c("integer", "integer", "double", "character"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_19040),
+ dim(output_17375),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_19040)),
+ colnames(dplyr::group_keys(output_17375)),
character(0),
fixed = TRUE)
@@ -1027,13 +1372,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_11387 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = NULL, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_18110 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = NULL, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_11387[['error']]),
+ xpectr::strip(side_effects_18110[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'shuffle_pairs': Must be of type 'logical flag', not 'NULL'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_11387[['error_class']]),
+ xpectr::strip(side_effects_18110[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1041,56 +1386,56 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: factor_name = ".pair"
xpectr::set_test_seed(42)
# Assigning output
- output_19888 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = ".pair", overwrite = TRUE)
+ output_13881 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = ".pair", overwrite = TRUE)
# Testing class
expect_equal(
- class(output_19888),
+ class(output_13881),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_19888[["index"]],
+ output_13881[["index"]],
c(1, 9, 2, 8, 5, 3, 7, 4, 6),
tolerance = 1e-4)
expect_equal(
- output_19888[["A"]],
+ output_13881[["A"]],
c(9, 2, 8, 1, 4, 3, 6, 5, 7),
tolerance = 1e-4)
expect_equal(
- output_19888[["B"]],
+ output_13881[["B"]],
c(0.70506, 0.11749, 0.45774, 0.97823, 0.25543, 0.71911, 0.94001,
0.93467, 0.46229),
tolerance = 1e-4)
expect_equal(
- output_19888[["C"]],
+ output_13881[["C"]],
c("A", "I", "B", "H", "E", "C", "G", "D", "F"),
fixed = TRUE)
expect_equal(
- output_19888[[".pair"]],
+ output_13881[[".pair"]],
structure(c(1L, 1L, 2L, 2L, 3L, 4L, 4L, 5L, 5L), .Label = c("1",
"2", "3", "4", "5"), class = "factor"))
# Testing column names
expect_equal(
- names(output_19888),
+ names(output_13881),
c("index", "A", "B", "C", ".pair"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_19888),
+ xpectr::element_classes(output_13881),
c("integer", "integer", "numeric", "character", "factor"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_19888),
+ xpectr::element_types(output_13881),
c("integer", "integer", "double", "character", "integer"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_19888),
+ dim(output_13881),
c(9L, 5L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_19888)),
+ colnames(dplyr::group_keys(output_13881)),
character(0),
fixed = TRUE)
@@ -1098,52 +1443,52 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: factor_name = "A"
xpectr::set_test_seed(42)
# Assigning output
- output_19466 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = TRUE)
+ output_16851 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = TRUE)
# Testing class
expect_equal(
- class(output_19466),
+ class(output_16851),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_19466[["index"]],
+ output_16851[["index"]],
c(1, 9, 2, 8, 5, 3, 7, 4, 6),
tolerance = 1e-4)
expect_equal(
- output_19466[["A"]],
- structure(c(1L, 1L, 2L, 2L, 3L, 4L, 4L, 5L, 5L), .Label = c("1",
- "2", "3", "4", "5"), class = "factor"))
- expect_equal(
- output_19466[["B"]],
+ output_16851[["B"]],
c(0.70506, 0.11749, 0.45774, 0.97823, 0.25543, 0.71911, 0.94001,
0.93467, 0.46229),
tolerance = 1e-4)
expect_equal(
- output_19466[["C"]],
+ output_16851[["C"]],
c("A", "I", "B", "H", "E", "C", "G", "D", "F"),
fixed = TRUE)
+ expect_equal(
+ output_16851[["A"]],
+ structure(c(1L, 1L, 2L, 2L, 3L, 4L, 4L, 5L, 5L), .Label = c("1",
+ "2", "3", "4", "5"), class = "factor"))
# Testing column names
expect_equal(
- names(output_19466),
- c("index", "A", "B", "C"),
+ names(output_16851),
+ c("index", "B", "C", "A"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_19466),
- c("integer", "factor", "numeric", "character"),
+ xpectr::element_classes(output_16851),
+ c("integer", "numeric", "character", "factor"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_19466),
- c("integer", "integer", "double", "character"),
+ xpectr::element_types(output_16851),
+ c("integer", "double", "character", "integer"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_19466),
+ dim(output_16851),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_19466)),
+ colnames(dplyr::group_keys(output_16851)),
character(0),
fixed = TRUE)
@@ -1152,13 +1497,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_10824 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = 1, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_10039 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = 1, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_10824[['error']]),
+ xpectr::strip(side_effects_10039[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'factor_name': Must be of type 'string' (or 'NULL'), not 'double'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_10824[['error_class']]),
+ xpectr::strip(side_effects_10039[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1167,13 +1512,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_15142 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NA, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_18329 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NA, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_15142[['error']]),
+ xpectr::strip(side_effects_18329[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'factor_name': May not be NA."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_15142[['error_class']]),
+ xpectr::strip(side_effects_18329[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1182,13 +1527,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_13902 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = FALSE), reset_seed = TRUE)
+ side_effects_10073 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = FALSE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_13902[['error']]),
- xpectr::strip("1 assertions failed:\n * The column 'A' already exists and 'overwrite' is disabled."),
+ xpectr::strip(side_effects_10073[['error']]),
+ xpectr::strip("Adding these dimensions would overwrite existing columns: A."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_13902[['error_class']]),
+ xpectr::strip(side_effects_10073[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1197,13 +1542,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_19057 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = NULL), reset_seed = TRUE)
+ side_effects_12076 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = NULL), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_19057[['error']]),
+ xpectr::strip(side_effects_12076[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'overwrite': Must be of type 'logical flag', not 'NULL'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_19057[['error_class']]),
+ xpectr::strip(side_effects_12076[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1233,8 +1578,7 @@ test_that("testing pair_extremes_vec()", {
# Testing values
expect_equal(
output_19148,
- c(0.64175, 0.83045, 0.28614, 0.93708, 0.91481, 0.5191, 0.65699,
- 0.73659, 0.13467),
+ c(0.91480, 0.5190959, 0.736588, 0.65699, 0.6417, 0.8304476, 0.134667, 0.2861395, 0.937075),
tolerance = 1e-4)
# Testing names
expect_equal(
@@ -1252,4 +1596,43 @@ test_that("testing pair_extremes_vec()", {
## Finished testing 'pair_extremes_vec(data = runif(9), unequal_m...' ####
+
+
+ ## Testing 'pair_extremes_vec(data = runif(9), unequal_m...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_16932 <- pair_extremes_vec(data = runif(9),
+ unequal_method = "first",
+ num_pairings = 2)
+ # Testing class
+ expect_equal(
+ class(output_16932),
+ "numeric",
+ fixed = TRUE)
+ # Testing type
+ expect_type(
+ output_16932,
+ type = "double")
+ # Testing values
+ expect_equal(
+ output_16932,
+ c(0.13467, 0.28614, 0.93708, 0.64175, 0.83045, 0.5191, 0.91481,
+ 0.65699, 0.73659),
+ tolerance = 1e-4)
+ # Testing names
+ expect_equal(
+ names(output_16932),
+ NULL,
+ fixed = TRUE)
+ # Testing length
+ expect_equal(
+ length(output_16932),
+ 9L)
+ # Testing sum of element lengths
+ expect_equal(
+ sum(xpectr::element_lengths(output_16932)),
+ 9L)
+ ## Finished testing 'pair_extremes_vec(data = runif(9), unequal_m...' ####
+
})
diff --git a/tests/testthat/test_shuffle_hierarchy.R b/tests/testthat/test_shuffle_hierarchy.R
new file mode 100644
index 0000000..50d36bb
--- /dev/null
+++ b/tests/testthat/test_shuffle_hierarchy.R
@@ -0,0 +1,593 @@
+library(rearrr)
+context("shuffle_hierarchy")
+
+
+test_that("shuffle_hierarchy()", {
+ xpectr::set_test_seed(42)
+
+ df <- data.frame(
+ 'a' = rep(1:2, each = 8),
+ 'b' = rep(1:4, each = 4),
+ 'c' = rep(1:8, each = 2),
+ 'd' = 1:16
+ )
+
+ ## Testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(98)
+ # Assigning output
+ output_19148 <- shuffle_hierarchy(df, group_cols = c('a', 'b', 'c', 'd'))
+ # Testing class
+ expect_equal(
+ class(output_19148),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19148[["a"]],
+ c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["b"]],
+ c(4L, 4L, 4L, 4L, 3L, 3L, 3L, 3L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["c"]],
+ c(8L, 8L, 7L, 7L, 6L, 6L, 5L, 5L, 1L, 1L, 2L, 2L, 4L, 4L, 3L, 3L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["d"]],
+ c(15L, 16L, 13L, 14L, 12L, 11L, 10L, 9L, 1L, 2L, 3L, 4L, 7L, 8L, 5L, 6L),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_19148),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19148),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19148)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+
+
+
+ ## Testing 'shuffle_hierarchy( data = df, group_cols = c...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(98)
+ # Assigning output
+ output_16988 <- shuffle_hierarchy(
+ data = df,
+ group_cols = c('a', 'b', 'c', 'd'),
+ cols_to_shuffle = c('a', 'c')
+ )
+ # Testing class
+ expect_equal(
+ class(output_16988),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_16988[["a"]],
+ c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_16988[["b"]],
+ c(3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_16988[["c"]],
+ c(5L, 5L, 6L, 6L, 7L, 7L, 8L, 8L, 1L, 1L, 2L, 2L, 3L, 3L, 4L, 4L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_16988[["d"]],
+ c(9L, 10L, 11L, 12L, 13L, 14L, 15L, 16L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_16988),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_16988),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_16988),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_16988),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_16988)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy( data = df, group_cols = c...' ####
+
+ ## Testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_10268 <- shuffle_hierarchy(df, group_cols = c('a', 'b'), leaf_has_groups = FALSE)
+ # Testing class
+ expect_equal(
+ class(output_10268),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_10268[["a"]],
+ c(2, 2, 2, 2, 2, 2, 2, 2, 1, 1, 1, 1, 1, 1, 1, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10268[["b"]],
+ c(4, 4, 3, 3, 4, 3, 4, 3, 2, 2, 1, 2, 1, 2, 1, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10268[["c"]],
+ c(7, 7, 6, 6, 8, 5, 8, 5, 4, 4, 1, 3, 2, 3, 2, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10268[["d"]],
+ c(14, 13, 11, 12, 15, 9, 16, 10, 8, 7, 2, 5, 3, 6, 4, 1),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_10268),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_10268),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_10268),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_10268),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_10268)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+
+ # If leaf col is not in cols_to_shuffle
+ # last column should be considered a group (because it is not the leaf)
+
+ ## Testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19782 <- shuffle_hierarchy(df, group_cols = c('a', 'b'), cols_to_shuffle = 'a', leaf_has_groups = FALSE)
+ # Testing class
+ expect_equal(
+ class(output_19782),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19782[["a"]],
+ c(2, 2, 2, 2, 2, 2, 2, 2, 1, 1, 1, 1, 1, 1, 1, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19782[["b"]],
+ c(3, 3, 3, 3, 4, 4, 4, 4, 1, 1, 1, 1, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19782[["c"]],
+ c(5, 5, 6, 6, 7, 7, 8, 8, 1, 1, 2, 2, 3, 3, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19782[["d"]],
+ c(9, 10, 11, 12, 13, 14, 15, 16, 1, 2, 3, 4, 5, 6, 7, 8),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_19782),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19782),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19782),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19782),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19782)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+
+})
+
+
+test_that("fuzz testing shuffle_hierarchy()", {
+ xpectr::set_test_seed(42)
+
+ df <- data.frame(
+ 'a' = rep(1:2, each = 8),
+ 'b' = rep(1:4, each = 4),
+ 'c' = rep(1:8, each = 2),
+ 'd' = 1:16
+ )
+
+ # Generate expectations for 'shuffle_hierarchy'
+ # Tip: comment out the gxs_function() call
+ # so it is easy to regenerate the tests
+ xpectr::set_test_seed(42)
+ # xpectr::gxs_function(
+ # fn = shuffle_hierarchy,
+ # args_values = list(
+ # "data" = list(df, c(1,2,3), NA),
+ # "group_cols" = list(c('a', 'b', 'c', 'd'), c(1, 2, 3, 4), NA),
+ # "cols_to_shuffle" = list(c('a', 'b', 'c', 'd'), c('b', 'c'), 2, NA),
+ # "leaf_has_groups" = list(TRUE, FALSE, NA)
+ # ),
+ # indentation = 2,
+ # copy_env = FALSE
+ # )
+
+ ## Testing 'shuffle_hierarchy' ####
+ ## Initially generated by xpectr
+ # Testing different combinations of argument values
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19148 <- shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_19148),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19148[["a"]],
+ c(1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["b"]],
+ c(1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["c"]],
+ c(2, 2, 1, 1, 3, 3, 4, 4, 6, 6, 5, 5, 8, 8, 7, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["d"]],
+ c(3, 4, 2, 1, 6, 5, 8, 7, 11, 12, 10, 9, 15, 16, 14, 13),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_19148),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19148),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19148)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = c(1, 2, 3), group_c...
+ # Changed from baseline: data = c(1, 2, 3)
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_19370 <- xpectr::capture_side_effects(shuffle_hierarchy(data = c(1, 2, 3), group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19370[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'data': Must be of type 'data.frame', not 'double'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19370[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = NA, group_cols = c(...
+ # Changed from baseline: data = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_12861 <- xpectr::capture_side_effects(shuffle_hierarchy(data = NA, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12861[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'data': Must be of type 'data.frame', not 'logical'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12861[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = NULL, group_cols = ...
+ # Changed from baseline: data = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_18304 <- xpectr::capture_side_effects(shuffle_hierarchy(data = NULL, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_18304[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'data': Must be of type 'data.frame', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_18304[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: group_cols = c(1, 2, ...
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_16417 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c(1, 2, 3, 4), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16417[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'group_cols': Must be of type 'character', not 'double'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16417[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = NA...
+ # Changed from baseline: group_cols = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_15190 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = NA, cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15190[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'group_cols': Contains missing values (element 1)."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15190[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = NU...
+ # Changed from baseline: group_cols = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_17365 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = NULL, cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17365[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'group_cols': Must be of type 'character', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17365[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = c("...
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_11346 <- shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("b", "c"), leaf_has_groups = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_11346),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_11346[["a"]],
+ c(1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11346[["b"]],
+ c(2, 2, 2, 2, 1, 1, 1, 1, 3, 3, 3, 3, 4, 4, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11346[["c"]],
+ c(3, 3, 4, 4, 2, 2, 1, 1, 6, 6, 5, 5, 8, 8, 7, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11346[["d"]],
+ c(5, 6, 7, 8, 3, 4, 1, 2, 11, 12, 9, 10, 15, 16, 13, 14),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_11346),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_11346),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_11346),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_11346),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_11346)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = 2
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_16569 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = 2, leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16569[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'cols_to_shuffle': Must be of type 'character', not 'double'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16569[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_17050 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = NA, leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17050[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'cols_to_shuffle': Contains missing values (element 1)."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17050[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_14577 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = NULL, leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_14577[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'cols_to_shuffle': Must be of type 'character', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_14577[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: leaf_has_groups = FALSE
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_17191 <- shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = FALSE)
+ # Testing class
+ expect_equal(
+ class(output_17191),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_17191[["a"]],
+ c(1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_17191[["b"]],
+ c(1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_17191[["c"]],
+ c(2, 2, 1, 1, 3, 3, 4, 4, 6, 6, 5, 5, 8, 8, 7, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_17191[["d"]],
+ c(3, 4, 2, 1, 6, 5, 8, 7, 11, 12, 10, 9, 15, 16, 14, 13),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_17191),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_17191),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_17191),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_17191),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_17191)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: leaf_has_groups = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_19346 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = NA), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19346[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'leaf_has_groups': May not be NA."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19346[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: leaf_has_groups = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_12554 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = NULL), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12554[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'leaf_has_groups': Must be of type 'logical flag', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12554[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ ## Finished testing 'shuffle_hierarchy' ####
+ #
+
+})
+
## Utilities
diff --git a/cran-comments.md b/cran-comments.md
index 4b4fe6a..02c18c2 100644
--- a/cran-comments.md
+++ b/cran-comments.md
@@ -5,9 +5,4 @@
## R CMD check results
-0 errors | 0 warnings | 1 note
-
-* This is a new release.
-* Removed examples for unexported functions
-* Made examples testable in most cases
-* Replaced <<- assignment with specific assignment to the right environment
+0 errors | 0 warnings | 0 notes
diff --git a/man/center_max.Rd b/man/center_max.Rd
index 729ce93..0a316f0 100644
--- a/man/center_max.Rd
+++ b/man/center_max.Rd
@@ -84,7 +84,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/center_min.Rd b/man/center_min.Rd
index e965bf7..0131e96 100644
--- a/man/center_min.Rd
+++ b/man/center_min.Rd
@@ -84,7 +84,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/closest_to.Rd b/man/closest_to.Rd
index 3c8a1d1..a585d95 100644
--- a/man/closest_to.Rd
+++ b/man/closest_to.Rd
@@ -165,7 +165,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
Other distance functions:
\code{\link{dim_values}()},
diff --git a/man/extreme_pairing_rearranger_.Rd b/man/extreme_pairing_rearranger_.Rd
index cb83c7e..4ede22f 100644
--- a/man/extreme_pairing_rearranger_.Rd
+++ b/man/extreme_pairing_rearranger_.Rd
@@ -11,6 +11,7 @@ extreme_pairing_rearranger_(
shuffle_members = FALSE,
shuffle_pairs = FALSE,
num_pairings = 1,
+ balance = "mean",
factor_name = ".pair",
overwrite = FALSE
)
@@ -31,7 +32,7 @@ The first group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -50,7 +51,7 @@ The middle group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -68,7 +69,7 @@ The last group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -86,15 +87,40 @@ And are \strong{ordered as}:
\item{shuffle_pairs}{Whether to shuffle the pairs. (Logical)}
+\item{num_pairings}{Number of pairings to perform (recursively). At least \code{1}.
+
+Based on \code{`balance`}, the secondary pairings perform extreme pairing on either the
+\emph{sum}, \emph{absolute difference}, \emph{min}, or \emph{max} of the pair elements.}
+
+\item{balance}{What to balance pairs for in a given \emph{secondary} pairing.
+Either \code{"mean"}, \code{"spread"}, \code{"min"}, or \code{"max"}.
+Can be a single string used for all secondary pairings
+or one for each secondary pairing (\code{`num_pairings` - 1}).
+
+The first pairing always pairs the actual element values.
+
+\subsection{mean}{
+Pairs have similar means. The values in the pairs from the previous pairing
+are aggregated with \code{`sum()`} and paired.
+}
+\subsection{spread}{
+Pairs have similar spread (e.g. standard deviations). The values in the pairs from the previous pairing
+are aggregated with \code{`sum(abs(diff()))`} and paired.
+}
+\subsection{min / max}{
+Pairs have similar minimum / maximum values. The values in the pairs from the previous pairing
+are aggregated with \code{`min()`} / \code{`max()`} and paired.
+}}
+
\item{factor_name}{Name of new column with the sorting factor. If \code{NULL}, no column is added.}
\item{overwrite}{Whether to allow overwriting of existing columns. (Logical)}
}
\value{
The sorted \code{data.frame} (\code{tibble}) / \code{vector}.
-Optionally with the sorting factor added.
+Optionally with the sorting factor(s) added.
-When \code{`data`} is a \code{vector} and \code{`keep_factors`} is \code{FALSE},
+When \code{`data`} is a \code{vector} and \code{`factor_name`} is \code{NULL},
the output will be a \code{vector}. Otherwise, a \code{data.frame}.
}
\description{
diff --git a/man/figures/README-unnamed-chunk-19-1.png b/man/figures/README-unnamed-chunk-19-1.png
new file mode 100644
index 0000000..e6e93ee
Binary files /dev/null and b/man/figures/README-unnamed-chunk-19-1.png differ
diff --git a/man/figures/README-unnamed-chunk-21-1.png b/man/figures/README-unnamed-chunk-21-1.png
new file mode 100644
index 0000000..766574a
Binary files /dev/null and b/man/figures/README-unnamed-chunk-21-1.png differ
diff --git a/man/figures/README-unnamed-chunk-27-1.png b/man/figures/README-unnamed-chunk-27-1.png
new file mode 100644
index 0000000..927a9f9
Binary files /dev/null and b/man/figures/README-unnamed-chunk-27-1.png differ
diff --git a/man/figures/README-unnamed-chunk-29-1.png b/man/figures/README-unnamed-chunk-29-1.png
new file mode 100644
index 0000000..3c5a2ff
Binary files /dev/null and b/man/figures/README-unnamed-chunk-29-1.png differ
diff --git a/man/figures/README-unnamed-chunk-31-1.png b/man/figures/README-unnamed-chunk-31-1.png
new file mode 100644
index 0000000..5e97abd
Binary files /dev/null and b/man/figures/README-unnamed-chunk-31-1.png differ
diff --git a/man/figures/README-unnamed-chunk-33-1.png b/man/figures/README-unnamed-chunk-33-1.png
new file mode 100644
index 0000000..2487964
Binary files /dev/null and b/man/figures/README-unnamed-chunk-33-1.png differ
diff --git a/man/figures/README-unnamed-chunk-35-1.png b/man/figures/README-unnamed-chunk-35-1.png
new file mode 100644
index 0000000..0e4b3d0
Binary files /dev/null and b/man/figures/README-unnamed-chunk-35-1.png differ
diff --git a/man/figures/README-unnamed-chunk-46-1.png b/man/figures/README-unnamed-chunk-46-1.png
new file mode 100644
index 0000000..24b9de6
Binary files /dev/null and b/man/figures/README-unnamed-chunk-46-1.png differ
diff --git a/man/furthest_from.Rd b/man/furthest_from.Rd
index 4f7980a..17afb31 100644
--- a/man/furthest_from.Rd
+++ b/man/furthest_from.Rd
@@ -166,7 +166,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
Other distance functions:
\code{\link{closest_to}()},
diff --git a/man/pair_extremes.Rd b/man/pair_extremes.Rd
index b0eb76b..08fe718 100644
--- a/man/pair_extremes.Rd
+++ b/man/pair_extremes.Rd
@@ -9,15 +9,19 @@ pair_extremes(
data,
col = NULL,
unequal_method = "middle",
+ num_pairings = 1,
+ balance = "mean",
shuffle_members = FALSE,
shuffle_pairs = FALSE,
- factor_name = ".pair",
+ factor_name = ifelse(num_pairings == 1, ".pair", ".pairing"),
overwrite = FALSE
)
pair_extremes_vec(
data,
unequal_method = "middle",
+ num_pairings = 1,
+ balance = "mean",
shuffle_members = FALSE,
shuffle_pairs = FALSE
)
@@ -38,7 +42,7 @@ The first group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -57,7 +61,7 @@ The middle group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -75,7 +79,7 @@ The last group will have size \code{1}.
\strong{Example}:
-The column values:
+The ordered column values:
\code{c(1, 2, 3, 4, 5)}
@@ -89,6 +93,31 @@ And are \strong{ordered as}:
}}
+\item{num_pairings}{Number of pairings to perform (recursively). At least \code{1}.
+
+Based on \code{`balance`}, the secondary pairings perform extreme pairing on either the
+\emph{sum}, \emph{absolute difference}, \emph{min}, or \emph{max} of the pair elements.}
+
+\item{balance}{What to balance pairs for in a given \emph{secondary} pairing.
+Either \code{"mean"}, \code{"spread"}, \code{"min"}, or \code{"max"}.
+Can be a single string used for all secondary pairings
+or one for each secondary pairing (\code{`num_pairings` - 1}).
+
+The first pairing always pairs the actual element values.
+
+\subsection{mean}{
+Pairs have similar means. The values in the pairs from the previous pairing
+are aggregated with \code{`sum()`} and paired.
+}
+\subsection{spread}{
+Pairs have similar spread (e.g. standard deviations). The values in the pairs from the previous pairing
+are aggregated with \code{`sum(abs(diff()))`} and paired.
+}
+\subsection{min / max}{
+Pairs have similar minimum / maximum values. The values in the pairs from the previous pairing
+are aggregated with \code{`min()`} / \code{`max()`} and paired.
+}}
+
\item{shuffle_members}{Whether to shuffle the pair members. (Logical)}
\item{shuffle_pairs}{Whether to shuffle the pairs. (Logical)}
@@ -166,6 +195,9 @@ pair_extremes(df, col = "A", shuffle_members = TRUE)
# Shuffle the order of the pairs
pair_extremes(df, col = "A", shuffle_pairs = TRUE)
+# Use recursive pairing
+pair_extremes(df, col = "A", num_pairings = 2)
+
# Grouped by G
df \%>\%
dplyr::select(G, A) \%>\% # For clarity
@@ -197,7 +229,8 @@ Other rearrange functions:
\code{\link{position_max}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/position_max.Rd b/man/position_max.Rd
index 751e6ea..06a8d0c 100644
--- a/man/position_max.Rd
+++ b/man/position_max.Rd
@@ -89,7 +89,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_min}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/position_min.Rd b/man/position_min.Rd
index f1e85ae..e12c003 100644
--- a/man/position_min.Rd
+++ b/man/position_min.Rd
@@ -91,7 +91,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_max}()},
\code{\link{rev_windows}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/rev_windows.Rd b/man/rev_windows.Rd
index a81f40b..59d46ae 100644
--- a/man/rev_windows.Rd
+++ b/man/rev_windows.Rd
@@ -91,7 +91,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_max}()},
\code{\link{position_min}()},
-\code{\link{roll_elements}()}
+\code{\link{roll_elements}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/roll_elements.Rd b/man/roll_elements.Rd
index f4c6bd8..e2ef28f 100644
--- a/man/roll_elements.Rd
+++ b/man/roll_elements.Rd
@@ -137,7 +137,8 @@ Other rearrange functions:
\code{\link{pair_extremes}()},
\code{\link{position_max}()},
\code{\link{position_min}()},
-\code{\link{rev_windows}()}
+\code{\link{rev_windows}()},
+\code{\link{shuffle_hierarchy}()}
}
\author{
Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
diff --git a/man/shuffle_hierarchy.Rd b/man/shuffle_hierarchy.Rd
new file mode 100644
index 0000000..dd49c84
--- /dev/null
+++ b/man/shuffle_hierarchy.Rd
@@ -0,0 +1,84 @@
+% Generated by roxygen2: do not edit by hand
+% Please edit documentation in R/shuffle_hierarchy.R
+\name{shuffle_hierarchy}
+\alias{shuffle_hierarchy}
+\title{Shuffle multi-column hierarchy of groups}
+\usage{
+shuffle_hierarchy(
+ data,
+ group_cols,
+ cols_to_shuffle = group_cols,
+ leaf_has_groups = TRUE
+)
+}
+\arguments{
+\item{data}{\code{data.frame}.}
+
+\item{group_cols}{Names of columns making up the group hierarchy.
+The last column is the \emph{leaf} and is shuffled first (if also in \code{`cols_to_shuffle`}).}
+
+\item{cols_to_shuffle}{Names of columns to shuffle hierarchically.
+By default, all the \code{`group_cols`} are shuffled.}
+
+\item{leaf_has_groups}{Whether the leaf column contains groups or values. (Logical)
+
+When the elements are \emph{group identifiers}, they are ordered sequentially and shuffled together.
+
+When the elements are \emph{values}, they are simply shuffled.}
+}
+\value{
+The shuffled \code{data.frame} (\code{tibble}).
+}
+\description{
+\Sexpr[results=rd, stage=render]{lifecycle::badge("experimental")}
+
+Shuffles a tree/hierarchy of groups, one column at a time.
+The levels in the last ("leaf") column are shuffled first, then the second-last column, and so on.
+Elements of the same group are ordered sequentially.
+}
+\examples{
+# Attach packages
+library(rearrr)
+library(dplyr)
+
+df <- data.frame(
+ 'a' = rep(1:4, each = 4),
+ 'b' = rep(1:8, each = 2),
+ 'c' = 1:16
+)
+
+# Set seed for reproducibility
+set.seed(2)
+
+# Shuffle all columns
+shuffle_hierarchy(df, group_cols = c('a', 'b', 'c'))
+
+# Don't shuffle 'b' but keep grouping by it
+# So 'c' will be shuffled within each group in 'b'
+shuffle_hierarchy(
+ data = df,
+ group_cols = c('a', 'b', 'c'),
+ cols_to_shuffle = c('a', 'c')
+)
+
+# Shuffle 'b' as if it's not a group column
+# so elements are independent within their group
+# (i.e. same-valued elements are not necessarily ordered sequentially)
+shuffle_hierarchy(df, group_cols = c('a', 'b'), leaf_has_groups = FALSE)
+}
+\seealso{
+Other rearrange functions:
+\code{\link{center_max}()},
+\code{\link{center_min}()},
+\code{\link{closest_to}()},
+\code{\link{furthest_from}()},
+\code{\link{pair_extremes}()},
+\code{\link{position_max}()},
+\code{\link{position_min}()},
+\code{\link{rev_windows}()},
+\code{\link{roll_elements}()}
+}
+\author{
+Ludvig Renbo Olsen, \email{r-pkgs@ludvigolsen.dk}
+}
+\concept{rearrange functions}
diff --git a/tests/testthat/test_pair_extremes.R b/tests/testthat/test_pair_extremes.R
index dda4aa2..52c915f 100644
--- a/tests/testthat/test_pair_extremes.R
+++ b/tests/testthat/test_pair_extremes.R
@@ -2,7 +2,7 @@ library(rearrr)
context("pair_extremes()")
-test_that("rearrange() with method pair_extremes works", {
+test_that("pair_extremes() works", {
# Create data frame
xpectr::set_test_seed(1)
@@ -70,7 +70,7 @@ test_that("rearrange() with method pair_extremes works", {
expect_equal(df_rearranged$score, c(69, 140, 79, 92, 85, 87))
})
-test_that("rearrange() with method pair_extremes throws expected errors", {
+test_that("pair_extremes() throws expected errors", {
df <- data.frame("a" = c(1,2,3))
## Testing 'rearrange(df, method = "pair_extremes", uneq...' ####
@@ -209,7 +209,9 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# "data" = list(df, head(df, 8), c(1,2,3,4,5,6,7), c(1,2,3,4),
# factor(c(1,2,3,4,5,6,7,1,2,3)), list(1,2,3), NA, 1),
# "col" = list(NULL, "A", "B", "C"),
- # "unequal_method" = list("middle", "first", "last"),
+ # "unequal_method" = list("middle", "first", "last", NA),
+ # "num_pairings" = list(1, 2, NA),
+ # "balance" = list("mean", "spread", NA),
# "shuffle_members" = list(FALSE, TRUE),
# "shuffle_pairs" = list(FALSE, TRUE),
# "factor_name" = list(NULL, ".pair", "A", 1, NA),
@@ -219,7 +221,10 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# list("data" = c(1,2,3,4), "col" = "A"),
# list("data" = c(1,2,3,4), "factor_name" = "another_name"),
# list("shuffle_members" = TRUE, "shuffle_pairs" = TRUE),
- # list("factor_name" = "A", "overwrite" = FALSE)
+ # list("factor_name" = "A", "overwrite" = FALSE),
+ # list("num_pairings" = 2, "balance" = "spread"),
+ # list("num_pairings" = 2, "balance" = "max"),
+ # list("num_pairings" = 2, "balance" = "min")
# ),
# indentation = 2
# )
@@ -232,7 +237,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Testing pair_extremes(data = df, col = NULL, unequal...
xpectr::set_test_seed(42)
# Assigning output
- output_19148 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19148 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19148),
@@ -285,7 +290,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = head(df, 8)
xpectr::set_test_seed(42)
# Assigning output
- output_19370 <- pair_extremes(data = head(df, 8), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19370 <- pair_extremes(data = head(df, 8), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19370),
@@ -338,7 +343,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = c(1, 2, 3, 4, ...
xpectr::set_test_seed(42)
# Assigning output
- output_12861 <- pair_extremes(data = c(1, 2, 3, 4, 5, 6, 7), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_12861 <- pair_extremes(data = c(1, 2, 3, 4, 5, 6, 7), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_12861),
@@ -371,7 +376,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = c(1, 2, 3, 4)
xpectr::set_test_seed(42)
# Assigning output
- output_18304 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_18304 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_18304),
@@ -404,7 +409,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = factor(c(1, 2,...
xpectr::set_test_seed(42)
# Assigning output
- output_16417 <- pair_extremes(data = factor(c(1, 2, 3, 4, 5, 6, 7, 1, 2, 3)), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_16417 <- pair_extremes(data = factor(c(1, 2, 3, 4, 5, 6, 7, 1, 2, 3)), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing is factor
expect_true(
is.factor(output_16417))
@@ -437,7 +442,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_15190 <- xpectr::capture_side_effects(pair_extremes(data = list(1, 2, 3), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_15190 <- xpectr::capture_side_effects(pair_extremes(data = list(1, 2, 3), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_15190[['error']]),
xpectr::strip("1 assertions failed:\n * when 'data' is not a data.frame, it cannot be a list."),
@@ -452,7 +457,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_17365 <- xpectr::capture_side_effects(pair_extremes(data = NA, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_17365 <- xpectr::capture_side_effects(pair_extremes(data = NA, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_17365[['error']]),
xpectr::strip("Assertion failed. One of the following must apply:\n * checkmate::check_data_frame(data): Must be of type 'data.frame', not 'logical'\n * checkmate::check_vector(data): Contains missing values (element 1)\n * checkmate::check_factor(data): Contains missing values (element 1)"),
@@ -466,7 +471,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data = 1
xpectr::set_test_seed(42)
# Assigning output
- output_11346 <- pair_extremes(data = 1, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_11346 <- pair_extremes(data = 1, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_11346),
@@ -500,7 +505,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_16569 <- xpectr::capture_side_effects(pair_extremes(data = NULL, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_16569 <- xpectr::capture_side_effects(pair_extremes(data = NULL, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_16569[['error']]),
xpectr::strip("Assertion failed. One of the following must apply:\n * checkmate::check_data_frame(data): Must be of type 'data.frame', not 'NULL'\n * checkmate::check_vector(data): Must be of type 'vector', not 'NULL'\n * checkmate::check_factor(data): Must be of type 'factor', not 'NULL'"),
@@ -515,7 +520,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_17050 <- xpectr::capture_side_effects(pair_extremes(data = c(1, 2, 3, 4), col = "A", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_17050 <- xpectr::capture_side_effects(pair_extremes(data = c(1, 2, 3, 4), col = "A", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_17050[['error']]),
xpectr::strip("1 assertions failed:\n * when 'data' is not a data.frame, 'col(s)' must be 'NULL'."),
@@ -529,7 +534,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: data, factor_name
xpectr::set_test_seed(42)
# Assigning output
- output_14577 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "another_name", overwrite = TRUE)
+ output_14577 <- pair_extremes(data = c(1, 2, 3, 4), col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "another_name", overwrite = TRUE)
# Testing class
expect_equal(
class(output_14577),
@@ -572,7 +577,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: col = "C"
xpectr::set_test_seed(42)
# Assigning output
- output_17191 <- pair_extremes(data = df, col = "C", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_17191 <- pair_extremes(data = df, col = "C", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_17191),
@@ -625,7 +630,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: col = "A"
xpectr::set_test_seed(42)
# Assigning output
- output_19346 <- pair_extremes(data = df, col = "A", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19346 <- pair_extremes(data = df, col = "A", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19346),
@@ -678,7 +683,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: col = "B"
xpectr::set_test_seed(42)
# Assigning output
- output_12554 <- pair_extremes(data = df, col = "B", unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_12554 <- pair_extremes(data = df, col = "B", unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_12554),
@@ -731,7 +736,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: unequal_method = "first"
xpectr::set_test_seed(42)
# Assigning output
- output_14622 <- pair_extremes(data = df, col = NULL, unequal_method = "first", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_14622 <- pair_extremes(data = df, col = NULL, unequal_method = "first", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_14622),
@@ -784,7 +789,7 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: unequal_method = "last"
xpectr::set_test_seed(42)
# Assigning output
- output_19400 <- pair_extremes(data = df, col = NULL, unequal_method = "last", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19400 <- pair_extremes(data = df, col = NULL, unequal_method = "last", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
class(output_19400),
@@ -834,70 +839,410 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
fixed = TRUE)
# Testing pair_extremes(data = df, col = NULL, unequal...
- # Changed from baseline: unequal_method = NULL
+ # Changed from baseline: unequal_method = NA
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_19782 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = NULL, shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_19782 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = NA, num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
xpectr::strip(side_effects_19782[['error']]),
- xpectr::strip("1 assertions failed:\n * Variable 'unequal_method': Must be of type 'string', not 'NULL'."),
+ xpectr::strip("1 assertions failed:\n * Variable 'unequal_method': May not be NA."),
fixed = TRUE)
expect_equal(
xpectr::strip(side_effects_19782[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: unequal_method = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_11174 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = NULL, num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_11174[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'unequal_method': Must be of type 'string', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_11174[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings = 2
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_14749 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_14749),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_14749[["index"]],
+ c(5, 4, 6, 2, 8, 1, 9, 3, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_14749[["A"]],
+ c(4, 5, 7, 8, 1, 9, 2, 3, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_14749[["B"]],
+ c(0.25543, 0.93467, 0.46229, 0.45774, 0.97823, 0.70506, 0.11749,
+ 0.71911, 0.94001),
+ tolerance = 1e-4)
+ expect_equal(
+ output_14749[["C"]],
+ c("E", "D", "F", "B", "H", "A", "I", "C", "G"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_14749),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_14749),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_14749),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_14749),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_14749)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_15603 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = NA, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15603[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'num_pairings': May not be NA."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15603[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_19040 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = NULL, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19040[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'num_pairings': Must be of type 'count', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19040[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings, balance
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_11387 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "spread", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_11387),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_11387[["index"]],
+ c(1, 9, 5, 3, 7, 2, 8, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11387[["A"]],
+ c(9, 2, 4, 3, 6, 8, 1, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11387[["B"]],
+ c(0.70506, 0.11749, 0.25543, 0.71911, 0.94001, 0.45774, 0.97823,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11387[["C"]],
+ c("A", "I", "E", "C", "G", "B", "H", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_11387),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_11387),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_11387),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_11387),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_11387)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings, balance
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19888 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "max", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_19888),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19888[["index"]],
+ c(1, 9, 5, 3, 7, 2, 8, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19888[["A"]],
+ c(9, 2, 4, 3, 6, 8, 1, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19888[["B"]],
+ c(0.70506, 0.11749, 0.25543, 0.71911, 0.94001, 0.45774, 0.97823,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19888[["C"]],
+ c("A", "I", "E", "C", "G", "B", "H", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_19888),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19888),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19888),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19888),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19888)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: num_pairings, balance
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19466 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 2, balance = "min", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_19466),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19466[["index"]],
+ c(1, 9, 5, 3, 7, 2, 8, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19466[["A"]],
+ c(9, 2, 4, 3, 6, 8, 1, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19466[["B"]],
+ c(0.70506, 0.11749, 0.25543, 0.71911, 0.94001, 0.45774, 0.97823,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19466[["C"]],
+ c("A", "I", "E", "C", "G", "B", "H", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_19466),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19466),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19466),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19466),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19466)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: balance = "spread"
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_10824 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "spread", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_10824),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_10824[["index"]],
+ c(1, 9, 2, 8, 5, 3, 7, 4, 6),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10824[["A"]],
+ c(9, 2, 8, 1, 4, 3, 6, 5, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10824[["B"]],
+ c(0.70506, 0.11749, 0.45774, 0.97823, 0.25543, 0.71911, 0.94001,
+ 0.93467, 0.46229),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10824[["C"]],
+ c("A", "I", "B", "H", "E", "C", "G", "D", "F"),
+ fixed = TRUE)
+ # Testing column names
+ expect_equal(
+ names(output_10824),
+ c("index", "A", "B", "C"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_10824),
+ c("integer", "integer", "numeric", "character"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_10824),
+ c("integer", "integer", "double", "character"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_10824),
+ c(9L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_10824)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: balance = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_15142 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = NA, shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15142[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'balance': Contains missing values (element 1)."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15142[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing pair_extremes(data = df, col = NULL, unequal...
+ # Changed from baseline: balance = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_13902 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = NULL, shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_13902[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'balance': Must be of type 'character', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_13902[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
# Testing pair_extremes(data = df, col = NULL, unequal...
# Changed from baseline: shuffle_members = TRUE
xpectr::set_test_seed(42)
# Assigning output
- output_11174 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = TRUE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
+ output_19057 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = TRUE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
- class(output_11174),
+ class(output_19057),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_11174[["index"]],
- c(9, 1, 8, 2, 5, 3, 7, 6, 4),
+ output_19057[["index"]],
+ c(6, 4, 2, 5, 8, 7, 3, 1, 9),
tolerance = 1e-4)
expect_equal(
- output_11174[["A"]],
- c(2, 9, 1, 8, 4, 3, 6, 7, 5),
+ output_19057[["A"]],
+ c(7, 5, 8, 4, 1, 6, 3, 9, 2),
tolerance = 1e-4)
expect_equal(
- output_11174[["B"]],
- c(0.11749, 0.70506, 0.97823, 0.45774, 0.25543, 0.71911, 0.94001,
- 0.46229, 0.93467),
+ output_19057[["B"]],
+ c(0.46229, 0.93467, 0.45774, 0.25543, 0.97823, 0.94001, 0.71911,
+ 0.70506, 0.11749),
tolerance = 1e-4)
expect_equal(
- output_11174[["C"]],
- c("I", "A", "H", "B", "E", "C", "G", "F", "D"),
+ output_19057[["C"]],
+ c("F", "D", "B", "E", "H", "G", "C", "A", "I"),
fixed = TRUE)
# Testing column names
expect_equal(
- names(output_11174),
+ names(output_19057),
c("index", "A", "B", "C"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_11174),
+ xpectr::element_classes(output_19057),
c("integer", "integer", "numeric", "character"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_11174),
+ xpectr::element_types(output_19057),
c("integer", "integer", "double", "character"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_11174),
+ dim(output_19057),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_11174)),
+ colnames(dplyr::group_keys(output_19057)),
character(0),
fixed = TRUE)
@@ -906,13 +1251,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_14749 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = NULL, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_14469 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = NULL, shuffle_pairs = FALSE, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_14749[['error']]),
+ xpectr::strip(side_effects_14469[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'shuffle_members': Must be of type 'logical flag', not 'NULL'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_14749[['error_class']]),
+ xpectr::strip(side_effects_14469[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -920,52 +1265,52 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: shuffle_members, shuf...
xpectr::set_test_seed(42)
# Assigning output
- output_15603 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = TRUE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
+ output_18360 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = TRUE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
- class(output_15603),
+ class(output_18360),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_15603[["index"]],
- c(5, 6, 4, 9, 1, 2, 8, 3, 7),
+ output_18360[["index"]],
+ c(7, 3, 2, 8, 5, 4, 6, 9, 1),
tolerance = 1e-4)
expect_equal(
- output_15603[["A"]],
- c(4, 7, 5, 2, 9, 8, 1, 3, 6),
+ output_18360[["A"]],
+ c(6, 3, 8, 1, 4, 5, 7, 2, 9),
tolerance = 1e-4)
expect_equal(
- output_15603[["B"]],
- c(0.25543, 0.46229, 0.93467, 0.11749, 0.70506, 0.45774, 0.97823,
- 0.71911, 0.94001),
+ output_18360[["B"]],
+ c(0.94001, 0.71911, 0.45774, 0.97823, 0.25543, 0.93467, 0.46229,
+ 0.11749, 0.70506),
tolerance = 1e-4)
expect_equal(
- output_15603[["C"]],
- c("E", "F", "D", "I", "A", "B", "H", "C", "G"),
+ output_18360[["C"]],
+ c("G", "C", "B", "H", "E", "D", "F", "I", "A"),
fixed = TRUE)
# Testing column names
expect_equal(
- names(output_15603),
+ names(output_18360),
c("index", "A", "B", "C"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_15603),
+ xpectr::element_classes(output_18360),
c("integer", "integer", "numeric", "character"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_15603),
+ xpectr::element_types(output_18360),
c("integer", "integer", "double", "character"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_15603),
+ dim(output_18360),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_15603)),
+ colnames(dplyr::group_keys(output_18360)),
character(0),
fixed = TRUE)
@@ -973,52 +1318,52 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: shuffle_pairs = TRUE
xpectr::set_test_seed(42)
# Assigning output
- output_19040 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
+ output_17375 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = TRUE, factor_name = NULL, overwrite = TRUE)
# Testing class
expect_equal(
- class(output_19040),
+ class(output_17375),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_19040[["index"]],
- c(5, 4, 6, 1, 9, 2, 8, 3, 7),
+ output_17375[["index"]],
+ c(9, 1, 2, 8, 5, 7, 3, 4, 6),
tolerance = 1e-4)
expect_equal(
- output_19040[["A"]],
- c(4, 5, 7, 9, 2, 8, 1, 3, 6),
+ output_17375[["A"]],
+ c(2, 9, 8, 1, 4, 6, 3, 5, 7),
tolerance = 1e-4)
expect_equal(
- output_19040[["B"]],
- c(0.25543, 0.93467, 0.46229, 0.70506, 0.11749, 0.45774, 0.97823,
- 0.71911, 0.94001),
+ output_17375[["B"]],
+ c(0.11749, 0.70506, 0.45774, 0.97823, 0.25543, 0.94001, 0.71911,
+ 0.93467, 0.46229),
tolerance = 1e-4)
expect_equal(
- output_19040[["C"]],
- c("E", "D", "F", "A", "I", "B", "H", "C", "G"),
+ output_17375[["C"]],
+ c("I", "A", "B", "H", "E", "G", "C", "D", "F"),
fixed = TRUE)
# Testing column names
expect_equal(
- names(output_19040),
+ names(output_17375),
c("index", "A", "B", "C"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_19040),
+ xpectr::element_classes(output_17375),
c("integer", "integer", "numeric", "character"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_19040),
+ xpectr::element_types(output_17375),
c("integer", "integer", "double", "character"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_19040),
+ dim(output_17375),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_19040)),
+ colnames(dplyr::group_keys(output_17375)),
character(0),
fixed = TRUE)
@@ -1027,13 +1372,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_11387 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = NULL, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_18110 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = NULL, factor_name = NULL, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_11387[['error']]),
+ xpectr::strip(side_effects_18110[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'shuffle_pairs': Must be of type 'logical flag', not 'NULL'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_11387[['error_class']]),
+ xpectr::strip(side_effects_18110[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1041,56 +1386,56 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: factor_name = ".pair"
xpectr::set_test_seed(42)
# Assigning output
- output_19888 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = ".pair", overwrite = TRUE)
+ output_13881 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = ".pair", overwrite = TRUE)
# Testing class
expect_equal(
- class(output_19888),
+ class(output_13881),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_19888[["index"]],
+ output_13881[["index"]],
c(1, 9, 2, 8, 5, 3, 7, 4, 6),
tolerance = 1e-4)
expect_equal(
- output_19888[["A"]],
+ output_13881[["A"]],
c(9, 2, 8, 1, 4, 3, 6, 5, 7),
tolerance = 1e-4)
expect_equal(
- output_19888[["B"]],
+ output_13881[["B"]],
c(0.70506, 0.11749, 0.45774, 0.97823, 0.25543, 0.71911, 0.94001,
0.93467, 0.46229),
tolerance = 1e-4)
expect_equal(
- output_19888[["C"]],
+ output_13881[["C"]],
c("A", "I", "B", "H", "E", "C", "G", "D", "F"),
fixed = TRUE)
expect_equal(
- output_19888[[".pair"]],
+ output_13881[[".pair"]],
structure(c(1L, 1L, 2L, 2L, 3L, 4L, 4L, 5L, 5L), .Label = c("1",
"2", "3", "4", "5"), class = "factor"))
# Testing column names
expect_equal(
- names(output_19888),
+ names(output_13881),
c("index", "A", "B", "C", ".pair"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_19888),
+ xpectr::element_classes(output_13881),
c("integer", "integer", "numeric", "character", "factor"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_19888),
+ xpectr::element_types(output_13881),
c("integer", "integer", "double", "character", "integer"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_19888),
+ dim(output_13881),
c(9L, 5L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_19888)),
+ colnames(dplyr::group_keys(output_13881)),
character(0),
fixed = TRUE)
@@ -1098,52 +1443,52 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
# Changed from baseline: factor_name = "A"
xpectr::set_test_seed(42)
# Assigning output
- output_19466 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = TRUE)
+ output_16851 <- pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = TRUE)
# Testing class
expect_equal(
- class(output_19466),
+ class(output_16851),
c("tbl_df", "tbl", "data.frame"),
fixed = TRUE)
# Testing column values
expect_equal(
- output_19466[["index"]],
+ output_16851[["index"]],
c(1, 9, 2, 8, 5, 3, 7, 4, 6),
tolerance = 1e-4)
expect_equal(
- output_19466[["A"]],
- structure(c(1L, 1L, 2L, 2L, 3L, 4L, 4L, 5L, 5L), .Label = c("1",
- "2", "3", "4", "5"), class = "factor"))
- expect_equal(
- output_19466[["B"]],
+ output_16851[["B"]],
c(0.70506, 0.11749, 0.45774, 0.97823, 0.25543, 0.71911, 0.94001,
0.93467, 0.46229),
tolerance = 1e-4)
expect_equal(
- output_19466[["C"]],
+ output_16851[["C"]],
c("A", "I", "B", "H", "E", "C", "G", "D", "F"),
fixed = TRUE)
+ expect_equal(
+ output_16851[["A"]],
+ structure(c(1L, 1L, 2L, 2L, 3L, 4L, 4L, 5L, 5L), .Label = c("1",
+ "2", "3", "4", "5"), class = "factor"))
# Testing column names
expect_equal(
- names(output_19466),
- c("index", "A", "B", "C"),
+ names(output_16851),
+ c("index", "B", "C", "A"),
fixed = TRUE)
# Testing column classes
expect_equal(
- xpectr::element_classes(output_19466),
- c("integer", "factor", "numeric", "character"),
+ xpectr::element_classes(output_16851),
+ c("integer", "numeric", "character", "factor"),
fixed = TRUE)
# Testing column types
expect_equal(
- xpectr::element_types(output_19466),
- c("integer", "integer", "double", "character"),
+ xpectr::element_types(output_16851),
+ c("integer", "double", "character", "integer"),
fixed = TRUE)
# Testing dimensions
expect_equal(
- dim(output_19466),
+ dim(output_16851),
c(9L, 4L))
# Testing group keys
expect_equal(
- colnames(dplyr::group_keys(output_19466)),
+ colnames(dplyr::group_keys(output_16851)),
character(0),
fixed = TRUE)
@@ -1152,13 +1497,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_10824 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = 1, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_10039 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = 1, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_10824[['error']]),
+ xpectr::strip(side_effects_10039[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'factor_name': Must be of type 'string' (or 'NULL'), not 'double'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_10824[['error_class']]),
+ xpectr::strip(side_effects_10039[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1167,13 +1512,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_15142 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NA, overwrite = TRUE), reset_seed = TRUE)
+ side_effects_18329 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NA, overwrite = TRUE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_15142[['error']]),
+ xpectr::strip(side_effects_18329[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'factor_name': May not be NA."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_15142[['error_class']]),
+ xpectr::strip(side_effects_18329[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1182,13 +1527,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_13902 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = FALSE), reset_seed = TRUE)
+ side_effects_10073 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = "A", overwrite = FALSE), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_13902[['error']]),
- xpectr::strip("1 assertions failed:\n * The column 'A' already exists and 'overwrite' is disabled."),
+ xpectr::strip(side_effects_10073[['error']]),
+ xpectr::strip("Adding these dimensions would overwrite existing columns: A."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_13902[['error_class']]),
+ xpectr::strip(side_effects_10073[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1197,13 +1542,13 @@ test_that("fuzz testing pair_extremes method for rearrange()", {
xpectr::set_test_seed(42)
# Testing side effects
# Assigning side effects
- side_effects_19057 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = NULL), reset_seed = TRUE)
+ side_effects_12076 <- xpectr::capture_side_effects(pair_extremes(data = df, col = NULL, unequal_method = "middle", num_pairings = 1, balance = "mean", shuffle_members = FALSE, shuffle_pairs = FALSE, factor_name = NULL, overwrite = NULL), reset_seed = TRUE)
expect_equal(
- xpectr::strip(side_effects_19057[['error']]),
+ xpectr::strip(side_effects_12076[['error']]),
xpectr::strip("1 assertions failed:\n * Variable 'overwrite': Must be of type 'logical flag', not 'NULL'."),
fixed = TRUE)
expect_equal(
- xpectr::strip(side_effects_19057[['error_class']]),
+ xpectr::strip(side_effects_12076[['error_class']]),
xpectr::strip(c("simpleError", "error", "condition")),
fixed = TRUE)
@@ -1233,8 +1578,7 @@ test_that("testing pair_extremes_vec()", {
# Testing values
expect_equal(
output_19148,
- c(0.64175, 0.83045, 0.28614, 0.93708, 0.91481, 0.5191, 0.65699,
- 0.73659, 0.13467),
+ c(0.91480, 0.5190959, 0.736588, 0.65699, 0.6417, 0.8304476, 0.134667, 0.2861395, 0.937075),
tolerance = 1e-4)
# Testing names
expect_equal(
@@ -1252,4 +1596,43 @@ test_that("testing pair_extremes_vec()", {
## Finished testing 'pair_extremes_vec(data = runif(9), unequal_m...' ####
+
+
+ ## Testing 'pair_extremes_vec(data = runif(9), unequal_m...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_16932 <- pair_extremes_vec(data = runif(9),
+ unequal_method = "first",
+ num_pairings = 2)
+ # Testing class
+ expect_equal(
+ class(output_16932),
+ "numeric",
+ fixed = TRUE)
+ # Testing type
+ expect_type(
+ output_16932,
+ type = "double")
+ # Testing values
+ expect_equal(
+ output_16932,
+ c(0.13467, 0.28614, 0.93708, 0.64175, 0.83045, 0.5191, 0.91481,
+ 0.65699, 0.73659),
+ tolerance = 1e-4)
+ # Testing names
+ expect_equal(
+ names(output_16932),
+ NULL,
+ fixed = TRUE)
+ # Testing length
+ expect_equal(
+ length(output_16932),
+ 9L)
+ # Testing sum of element lengths
+ expect_equal(
+ sum(xpectr::element_lengths(output_16932)),
+ 9L)
+ ## Finished testing 'pair_extremes_vec(data = runif(9), unequal_m...' ####
+
})
diff --git a/tests/testthat/test_shuffle_hierarchy.R b/tests/testthat/test_shuffle_hierarchy.R
new file mode 100644
index 0000000..50d36bb
--- /dev/null
+++ b/tests/testthat/test_shuffle_hierarchy.R
@@ -0,0 +1,593 @@
+library(rearrr)
+context("shuffle_hierarchy")
+
+
+test_that("shuffle_hierarchy()", {
+ xpectr::set_test_seed(42)
+
+ df <- data.frame(
+ 'a' = rep(1:2, each = 8),
+ 'b' = rep(1:4, each = 4),
+ 'c' = rep(1:8, each = 2),
+ 'd' = 1:16
+ )
+
+ ## Testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(98)
+ # Assigning output
+ output_19148 <- shuffle_hierarchy(df, group_cols = c('a', 'b', 'c', 'd'))
+ # Testing class
+ expect_equal(
+ class(output_19148),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19148[["a"]],
+ c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["b"]],
+ c(4L, 4L, 4L, 4L, 3L, 3L, 3L, 3L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["c"]],
+ c(8L, 8L, 7L, 7L, 6L, 6L, 5L, 5L, 1L, 1L, 2L, 2L, 4L, 4L, 3L, 3L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["d"]],
+ c(15L, 16L, 13L, 14L, 12L, 11L, 10L, 9L, 1L, 2L, 3L, 4L, 7L, 8L, 5L, 6L),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_19148),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19148),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19148)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+
+
+
+ ## Testing 'shuffle_hierarchy( data = df, group_cols = c...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(98)
+ # Assigning output
+ output_16988 <- shuffle_hierarchy(
+ data = df,
+ group_cols = c('a', 'b', 'c', 'd'),
+ cols_to_shuffle = c('a', 'c')
+ )
+ # Testing class
+ expect_equal(
+ class(output_16988),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_16988[["a"]],
+ c(2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_16988[["b"]],
+ c(3L, 3L, 3L, 3L, 4L, 4L, 4L, 4L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_16988[["c"]],
+ c(5L, 5L, 6L, 6L, 7L, 7L, 8L, 8L, 1L, 1L, 2L, 2L, 3L, 3L, 4L, 4L),
+ tolerance = 1e-4)
+ expect_equal(
+ output_16988[["d"]],
+ c(9L, 10L, 11L, 12L, 13L, 14L, 15L, 16L, 1L, 2L, 3L, 4L, 5L, 6L, 7L, 8L),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_16988),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_16988),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_16988),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_16988),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_16988)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy( data = df, group_cols = c...' ####
+
+ ## Testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_10268 <- shuffle_hierarchy(df, group_cols = c('a', 'b'), leaf_has_groups = FALSE)
+ # Testing class
+ expect_equal(
+ class(output_10268),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_10268[["a"]],
+ c(2, 2, 2, 2, 2, 2, 2, 2, 1, 1, 1, 1, 1, 1, 1, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10268[["b"]],
+ c(4, 4, 3, 3, 4, 3, 4, 3, 2, 2, 1, 2, 1, 2, 1, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10268[["c"]],
+ c(7, 7, 6, 6, 8, 5, 8, 5, 4, 4, 1, 3, 2, 3, 2, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_10268[["d"]],
+ c(14, 13, 11, 12, 15, 9, 16, 10, 8, 7, 2, 5, 3, 6, 4, 1),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_10268),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_10268),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_10268),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_10268),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_10268)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+
+ # If leaf col is not in cols_to_shuffle
+ # last column should be considered a group (because it is not the leaf)
+
+ ## Testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+ ## Initially generated by xpectr
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19782 <- shuffle_hierarchy(df, group_cols = c('a', 'b'), cols_to_shuffle = 'a', leaf_has_groups = FALSE)
+ # Testing class
+ expect_equal(
+ class(output_19782),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19782[["a"]],
+ c(2, 2, 2, 2, 2, 2, 2, 2, 1, 1, 1, 1, 1, 1, 1, 1),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19782[["b"]],
+ c(3, 3, 3, 3, 4, 4, 4, 4, 1, 1, 1, 1, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19782[["c"]],
+ c(5, 5, 6, 6, 7, 7, 8, 8, 1, 1, 2, 2, 3, 3, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19782[["d"]],
+ c(9, 10, 11, 12, 13, 14, 15, 16, 1, 2, 3, 4, 5, 6, 7, 8),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_19782),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19782),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19782),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19782),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19782)),
+ character(0),
+ fixed = TRUE)
+ ## Finished testing 'shuffle_hierarchy(df, group_cols = c('a', 'b...' ####
+
+})
+
+
+test_that("fuzz testing shuffle_hierarchy()", {
+ xpectr::set_test_seed(42)
+
+ df <- data.frame(
+ 'a' = rep(1:2, each = 8),
+ 'b' = rep(1:4, each = 4),
+ 'c' = rep(1:8, each = 2),
+ 'd' = 1:16
+ )
+
+ # Generate expectations for 'shuffle_hierarchy'
+ # Tip: comment out the gxs_function() call
+ # so it is easy to regenerate the tests
+ xpectr::set_test_seed(42)
+ # xpectr::gxs_function(
+ # fn = shuffle_hierarchy,
+ # args_values = list(
+ # "data" = list(df, c(1,2,3), NA),
+ # "group_cols" = list(c('a', 'b', 'c', 'd'), c(1, 2, 3, 4), NA),
+ # "cols_to_shuffle" = list(c('a', 'b', 'c', 'd'), c('b', 'c'), 2, NA),
+ # "leaf_has_groups" = list(TRUE, FALSE, NA)
+ # ),
+ # indentation = 2,
+ # copy_env = FALSE
+ # )
+
+ ## Testing 'shuffle_hierarchy' ####
+ ## Initially generated by xpectr
+ # Testing different combinations of argument values
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_19148 <- shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_19148),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_19148[["a"]],
+ c(1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["b"]],
+ c(1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["c"]],
+ c(2, 2, 1, 1, 3, 3, 4, 4, 6, 6, 5, 5, 8, 8, 7, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_19148[["d"]],
+ c(3, 4, 2, 1, 6, 5, 8, 7, 11, 12, 10, 9, 15, 16, 14, 13),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_19148),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_19148),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_19148),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_19148)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = c(1, 2, 3), group_c...
+ # Changed from baseline: data = c(1, 2, 3)
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_19370 <- xpectr::capture_side_effects(shuffle_hierarchy(data = c(1, 2, 3), group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19370[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'data': Must be of type 'data.frame', not 'double'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19370[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = NA, group_cols = c(...
+ # Changed from baseline: data = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_12861 <- xpectr::capture_side_effects(shuffle_hierarchy(data = NA, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12861[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'data': Must be of type 'data.frame', not 'logical'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12861[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = NULL, group_cols = ...
+ # Changed from baseline: data = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_18304 <- xpectr::capture_side_effects(shuffle_hierarchy(data = NULL, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_18304[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'data': Must be of type 'data.frame', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_18304[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: group_cols = c(1, 2, ...
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_16417 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c(1, 2, 3, 4), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16417[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'group_cols': Must be of type 'character', not 'double'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16417[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = NA...
+ # Changed from baseline: group_cols = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_15190 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = NA, cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15190[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'group_cols': Contains missing values (element 1)."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_15190[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = NU...
+ # Changed from baseline: group_cols = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_17365 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = NULL, cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17365[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'group_cols': Must be of type 'character', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17365[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = c("...
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_11346 <- shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("b", "c"), leaf_has_groups = TRUE)
+ # Testing class
+ expect_equal(
+ class(output_11346),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_11346[["a"]],
+ c(1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11346[["b"]],
+ c(2, 2, 2, 2, 1, 1, 1, 1, 3, 3, 3, 3, 4, 4, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11346[["c"]],
+ c(3, 3, 4, 4, 2, 2, 1, 1, 6, 6, 5, 5, 8, 8, 7, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_11346[["d"]],
+ c(5, 6, 7, 8, 3, 4, 1, 2, 11, 12, 9, 10, 15, 16, 13, 14),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_11346),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_11346),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_11346),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_11346),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_11346)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = 2
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_16569 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = 2, leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16569[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'cols_to_shuffle': Must be of type 'character', not 'double'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_16569[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_17050 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = NA, leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17050[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'cols_to_shuffle': Contains missing values (element 1)."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_17050[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: cols_to_shuffle = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_14577 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = NULL, leaf_has_groups = TRUE), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_14577[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'cols_to_shuffle': Must be of type 'character', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_14577[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: leaf_has_groups = FALSE
+ xpectr::set_test_seed(42)
+ # Assigning output
+ output_17191 <- shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = FALSE)
+ # Testing class
+ expect_equal(
+ class(output_17191),
+ c("tbl_df", "tbl", "data.frame"),
+ fixed = TRUE)
+ # Testing column values
+ expect_equal(
+ output_17191[["a"]],
+ c(1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 2, 2, 2, 2, 2, 2),
+ tolerance = 1e-4)
+ expect_equal(
+ output_17191[["b"]],
+ c(1, 1, 1, 1, 2, 2, 2, 2, 3, 3, 3, 3, 4, 4, 4, 4),
+ tolerance = 1e-4)
+ expect_equal(
+ output_17191[["c"]],
+ c(2, 2, 1, 1, 3, 3, 4, 4, 6, 6, 5, 5, 8, 8, 7, 7),
+ tolerance = 1e-4)
+ expect_equal(
+ output_17191[["d"]],
+ c(3, 4, 2, 1, 6, 5, 8, 7, 11, 12, 10, 9, 15, 16, 14, 13),
+ tolerance = 1e-4)
+ # Testing column names
+ expect_equal(
+ names(output_17191),
+ c("a", "b", "c", "d"),
+ fixed = TRUE)
+ # Testing column classes
+ expect_equal(
+ xpectr::element_classes(output_17191),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing column types
+ expect_equal(
+ xpectr::element_types(output_17191),
+ c("integer", "integer", "integer", "integer"),
+ fixed = TRUE)
+ # Testing dimensions
+ expect_equal(
+ dim(output_17191),
+ c(16L, 4L))
+ # Testing group keys
+ expect_equal(
+ colnames(dplyr::group_keys(output_17191)),
+ character(0),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: leaf_has_groups = NA
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_19346 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = NA), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19346[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'leaf_has_groups': May not be NA."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_19346[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ # Testing shuffle_hierarchy(data = df, group_cols = c(...
+ # Changed from baseline: leaf_has_groups = NULL
+ xpectr::set_test_seed(42)
+ # Testing side effects
+ # Assigning side effects
+ side_effects_12554 <- xpectr::capture_side_effects(shuffle_hierarchy(data = df, group_cols = c("a", "b", "c", "d"), cols_to_shuffle = c("a", "b", "c", "d"), leaf_has_groups = NULL), reset_seed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12554[['error']]),
+ xpectr::strip("1 assertions failed:\n * Variable 'leaf_has_groups': Must be of type 'logical flag', not 'NULL'."),
+ fixed = TRUE)
+ expect_equal(
+ xpectr::strip(side_effects_12554[['error_class']]),
+ xpectr::strip(c("simpleError", "error", "condition")),
+ fixed = TRUE)
+
+ ## Finished testing 'shuffle_hierarchy' ####
+ #
+
+})
+