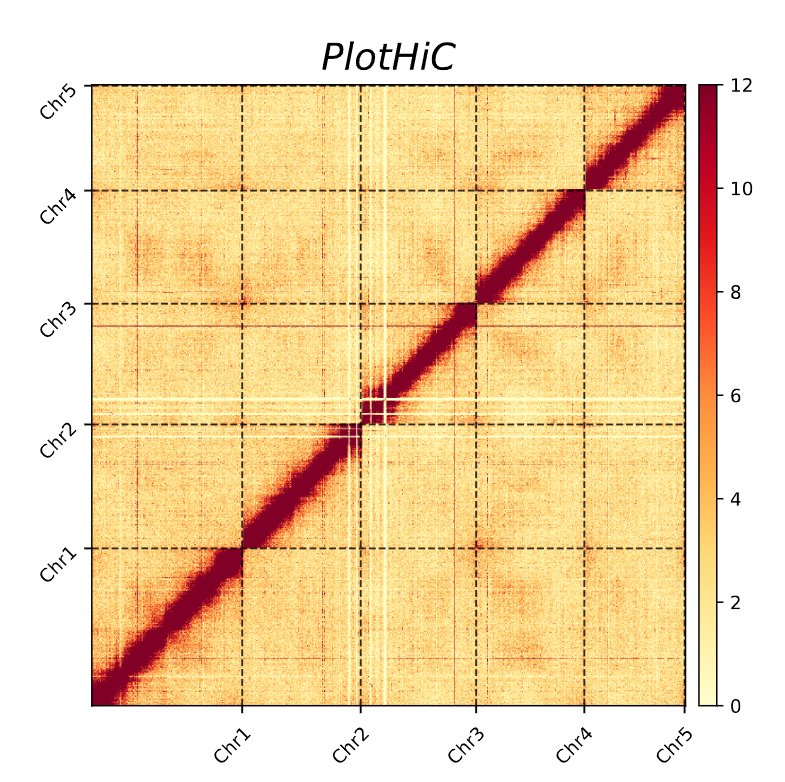
PlotHiC is an extension of AutoHiC that is used to visualize global interaction heatmaps after genome scaffolding.
Note: PlotHiC is currently under development. If you have any questions, please Open Issues or provide us with your comments via the email below.
Author: Zijie Jiang
Email: jzjlab@163.com
If you used PlotHiC in your research, please cite us:
Zijie Jiang, Zhixiang Peng, Zhaoyuan Wei, Jiahe Sun, Yongjiang Luo, Lingzi Bie, Guoqing Zhang, Yi Wang, A deep learning-based method enables the automatic and accurate assembly of chromosome-level genomes, Nucleic Acids Research, 2024;, gkae789, https://doi.org/10.1093/nar/gkae789
- Dependency :
python = "^3.10"
# pip install
pip install plothic
hic
This file is taken directly from 3d-dna, you need to select the final hic file (which has already been error adjusted and chromosome boundaries determined).
- Chromosome txt
This file is used for heatmap labeling. The first column is the name of the chromosome and the second column is the length of the chromosome (this length is the length of the hic file in Juicebox and can be manually determined from Juicebox).
Note: the length is in .hic file, not true base length.
# name length
Chr1 24800000
Chr2 44380000
Chr3 63338000
Chr4 81187000
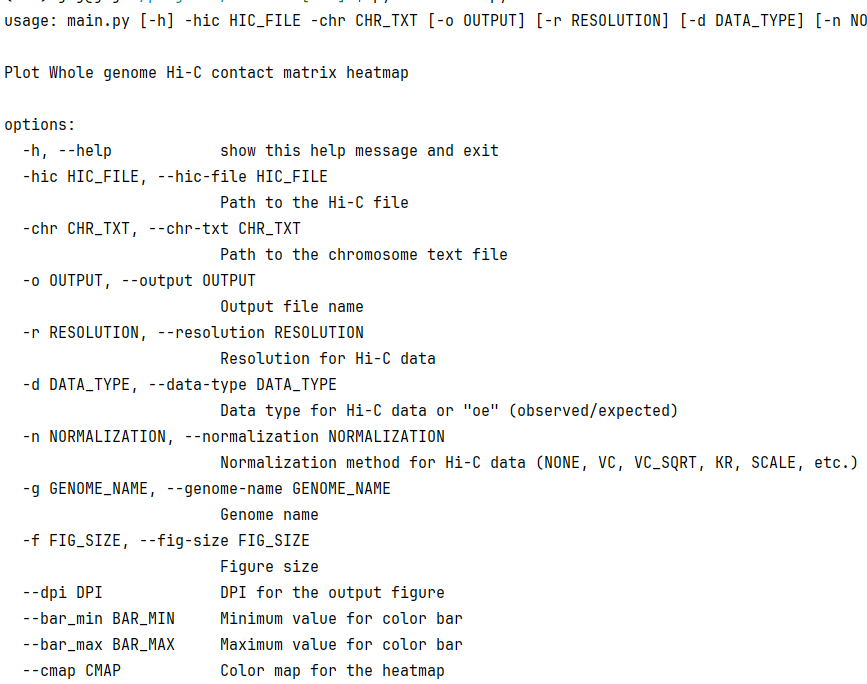
Chr5 97650000plothic -hic test.hic -chr chr.txt -r 100000
# -hic > .hic file
# -chr > chromosome length (in .hic file)
# -r > resolution to visualization
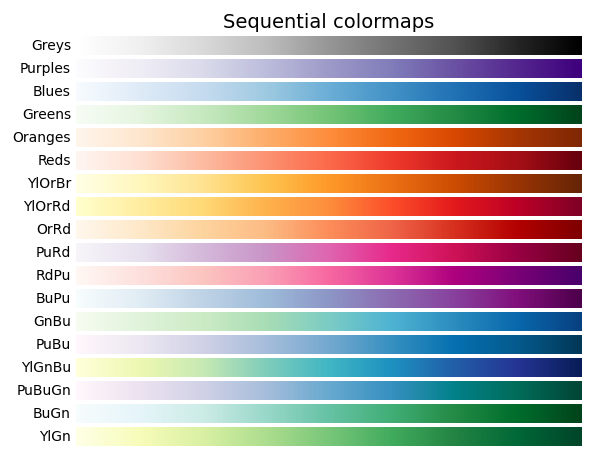
PlotHiC uses YlOrRd by default, you can choose more colors from Matplotlib.
Currently only use hic and chr txt are supported for visualization, and assembly files will be supported in the future. However, from the perspective of usage, using chr txt files seems to be more convenient. If you have better suggestions or other requirements, please Open Issues.