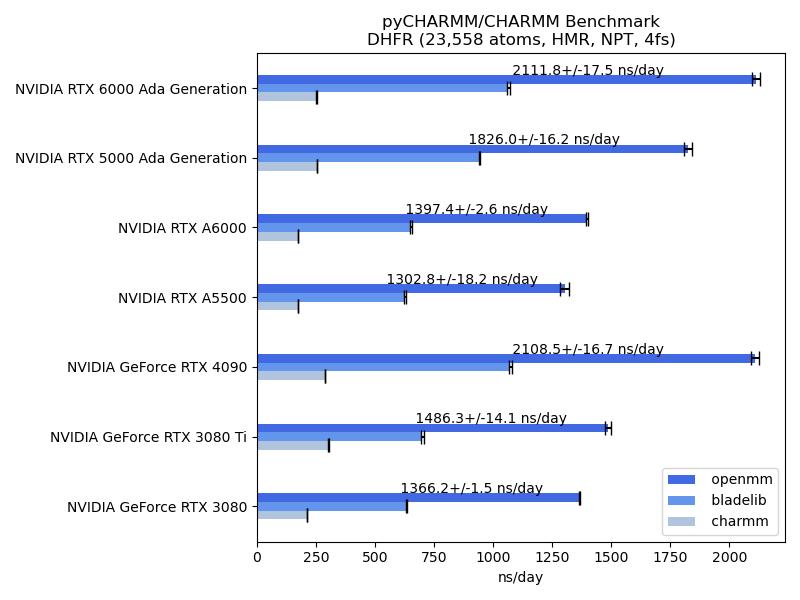
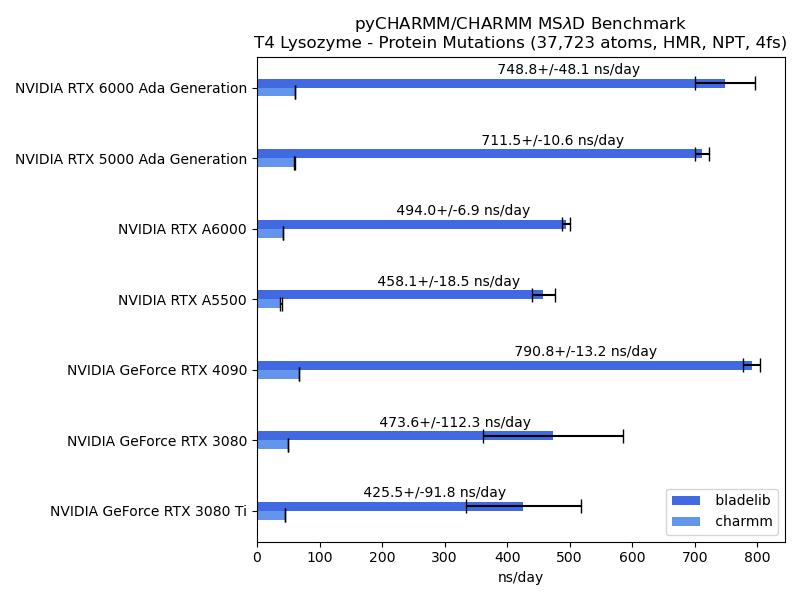
It is notable that CHARMM supports three GPU accelerated engines for molecular simulations, including free energy calcuations utilizing the highly scalable multi-site lambda-dynamics ($MS\lambda D$ ) framework for free energy calculations. CHARMM also provides a fully programmable interpreted language, from its inception in the late 1970s, and more recently a fully embedded Python API, pyCHARMM, that enables complex programs and workflows to be developed in Python and CHARMM scripting language.
This repository contains benchmarks for the CHARMM/pyCHARMM program for computations run on current GPU platforms. These include the standard JAC/DHFR, ApoA1, DMPG and STMV benchmarks, ranging in system size from ~23K atoms to ~1M atoms. In addition, new benchmarks illustrate the application of $MS\lambda D$ for both protein (T4 Lysoyme) and ligand (HSP90) relative free energy (RFE) calculations. The ApoA1, DMPG and STMV, as well as the two $MS\lambda D$ benchmarks have been updated to utilize the recent CHARMM 36 parameter sets. The JAC/DHFR benchmark is adapted from the early benchmark developed by Brooks and Case for CHARMM and Amber.
The benchmarks are all run from a single Python script at the top level of this repository, benchmark.py, and this set-up is mirrored in the Jupyter notebook benchmark.ipynb. An additional Jupyter notebook that can be utilized to plot the collected benchmarks resides in plot_benchmarks.ipynb.
The benchmarks were largely run on hardware present in the Brooks lab at the University of Michigan, with exception of the data for the RTX 4090, RTX 3080 TI and RTX 3080 which were generated by Parveen Gartan from Bergen University, Norway, on local hardware accessible to him.
All benchmarks were run in replicate to provide statistical variance of the resulting numbers through a slurm queueing system, and simple slurm job files are contained.
Multi-site ligand relative binding free energy calculations in HSP90 - hydrogen mass repartitioning (HMR), 4 fs, NPT
Multi-site protein stability changes from side chain mutations in T4 Lysozyme - hydrogen mass repartitioning (HMR), 4 fs, NPT
The benchmarks are all run from a single Python script
benchmark.pyand the timing results are appended to the created csv filebenchmark.csv.The data files, rtf, parameter, psf and cooordinate files are all stored in the benchmark subdirectories
<benchmark_name/engine_name/>(MD benchmarks) or<benchmark_name/engine_name/template>($MS\lambda D$ benchmarks), wherebenchmark_nameis 5_newDHFR, apoa1_bench, 6_dmpg, stmv_bench for the MD benchmarks and 2_T4L and 4_HSP90 for the$MS\lambda D$ benchmarks.engine_namerefers to openmm, bladelib or charmm (domdec) GPU accelerated engines.The parameters controlling the simulations are all stored in
yamlfilesbenchmark.ymlunder theengine_namesubdirectories.